Book traversal links for 2.6.2 Genotypic DST
Phenotypic DST remains the reference standard for most anti-TB compounds; however, this method is slow, and it requires specialized infrastructure and highly skilled staff. Genotypic DST (also referred to as molecular DST) holds promise to overcome some of these obstacles. Currently, available WHO-recommended rapid molecular drug susceptibility tests (Section 2.4) can be used to detect specific mutations known to confer phenotypic resistance. Although genotypic DST has advanced over the past decade, the spectrum of drugs covered is limited and does not cover the new and repurposed drug. However, rapid molecular tests for resistance to RIF, INH and FQ are feasible to implement in decentralized settings; such tests can deliver rapid results to inform initial treatment regimen selection while awaiting follow-on DST for other anti-TB drugs.
New and rapid next-generation technologies are thus needed to cover all priority drugs and to be accessible at the peripheral level, to expedite appropriate therapy and improve patient outcomes. WHO has developed a target product profile (TPP) to guide research and development to address this need (28).
DNA sequencing using NGS technologies is a promising method for the rapid detection of mutations associated with drug resistance for many anti-TB drugs (29). NGS-based DST could reduce the need for phenotypic DST for patient-care decisions, and it may be particularly useful for drugs for which phenotypic DST is unreliable or in settings that do not have the capacity to perform phenotypic DST.
NGS refers to techniques that rely on the sequencing of multiple DNA fragments in parallel, followed by bioinformatics analyses to assemble the sequences. The technologies can be used to determine the nucleotide sequence of an entire genome (i.e. WGS) or part of a genome (i.e. targeted NGS) in a single sequencing run. WGS and targeted NGS are recommended for use in the surveillance of DR-TB. WGS is used on cultured MTBC isolates whereas targeted NGS can be used directly on sputum specimens (30). In Section 2.4.4, the targeted NGS tests for the detection of resistance to anti-TB medicines recently recommended by WHO are described for use directly on clinical samples. These tests can detect mutations associated with resistance to RIF, INH, PZA, EMB, FQ, BDQ, LZD, CFZ, AMK and STR.
One important issue identified when using NGS tests was the lack of a standardized and robust single reference source for the interpretation of mutations. To address this need, WHO developed guidance and released the second edition of the catalogue of mutations in MTBC and their association with resistance, in 2023 (4). The catalogue provides a reference standard for the interpretation of mutations conferring resistance to all first-line and a variety of second-line drugs. The report summarizes the analysis of over 52 000 isolates with matched data on WGS and phenotypic DST from 67 countries for 13 anti-TB medicines. The catalogue provides information on more than 30 000 mutations, along with their frequencies within the dataset. The mutations are categorized into groups: those associated with resistance, those not associated with resistance, and those with uncertain significance (a significant number of mutations are in this category). Additionally, the catalogue provides details about the methods employed and important findings related to each drug. Future regular updates of the catalogue are planned.
Importantly, a transition to rapid molecular testing does not eliminate the need for phenotypic DST because the method is still needed for conducting DST for drugs for which a molecular tool is not available or for which resistance has not been clearly associated with specific mutations, conducting DST to guide drug dosing determinations, monitoring the response to TB treatment, and investigating discordant results from diagnostic testing or DST. In particular, phenotypic DST is needed for testing some of the new and repurposed drugs used for treatment; especially important in this context are BDQ, LZD and Pa.
Table 2.4 presents an overview of the WHO-recommended diagnostic approaches, reference methods and clinical interpretation for anti-TB medicines.
Table 2.4. WHO-recommended diagnostic approaches, reference methods and clinical interpretation for anti-TB medicines
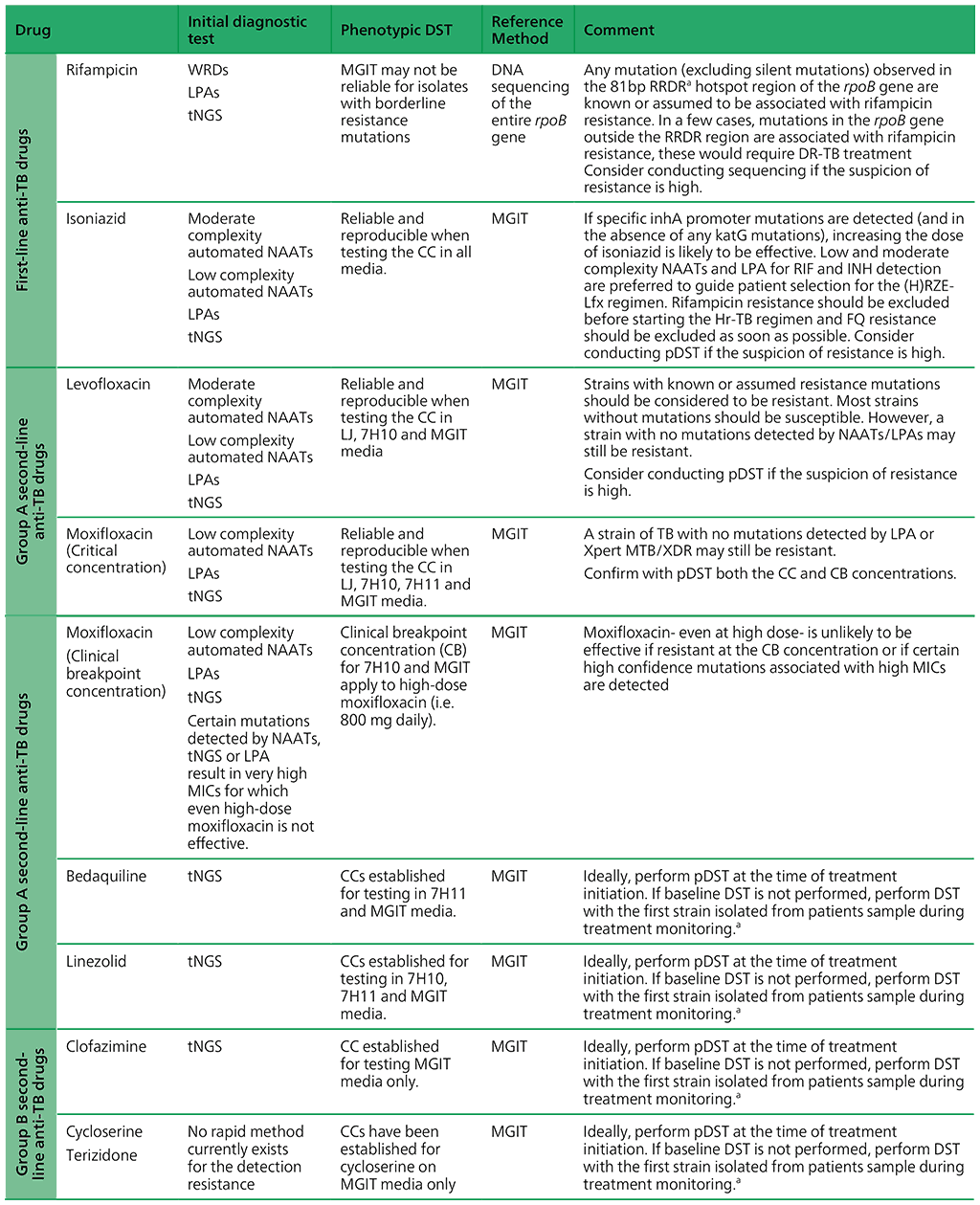
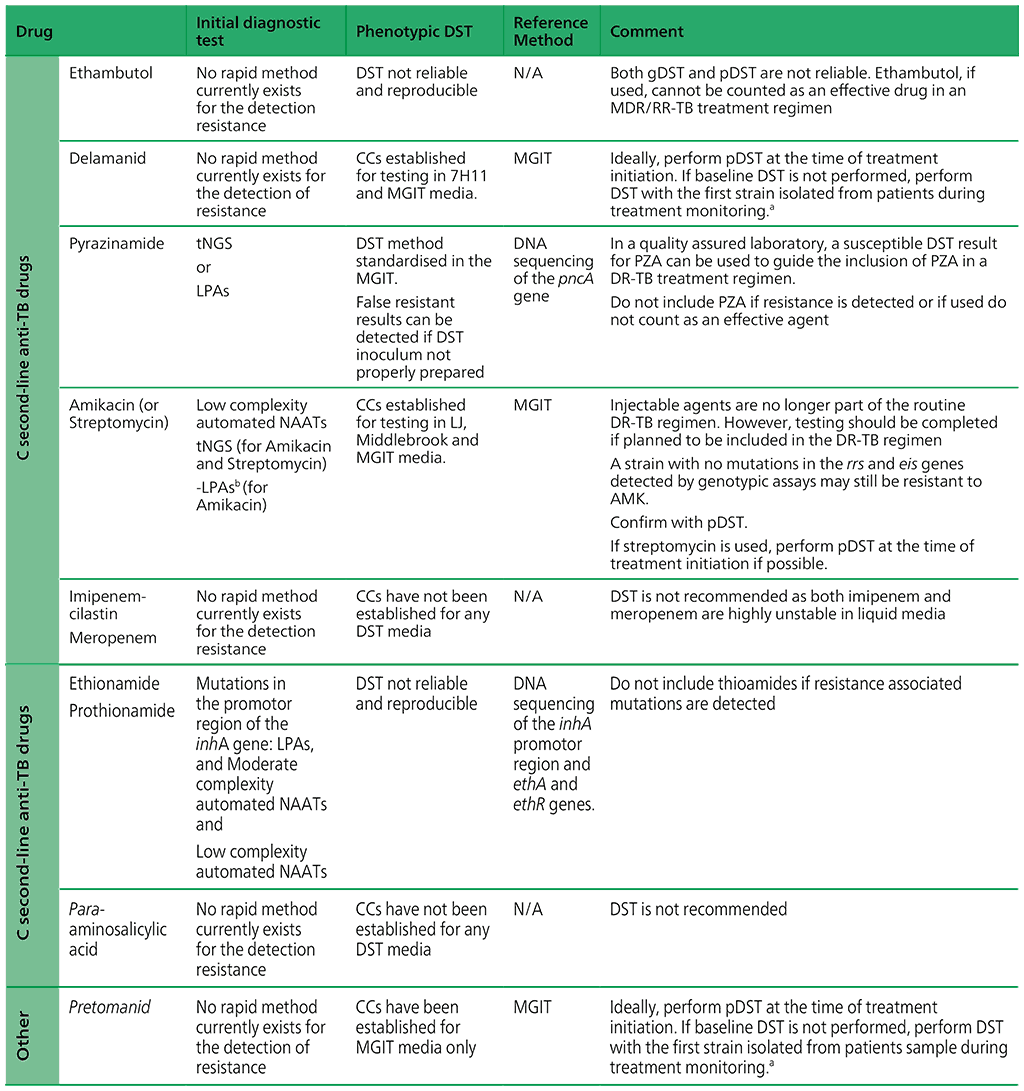
AMK: amikacin; ATU: area of technical uncertainty; CB: clinical breakpoint; CC: critical concentration; DNA: deoxyribonucleic acid; DR-TB: drug-resistant TB; DST: drug susceptibility testing; EMB: ethambutol; FQ: fluoroquinolone; Hr-TB: isoniazid-resistant, rifampicin-susceptible TB; HREZ: isoniazid (H), rifampicin (R), ethambutol (E) and pyrazinamide (Z); INH: isoniazid; LFX: levofloxacin; LJ: Löwenstein– Jensen media; LPA: line probe assay; MFX: moxifloxacin; MGIT: Mycobacterial Growth Indicator Tube; MIC: minimal inhibitory concentration; n.a: not available; NAAT: nucleic acid amplification test; NGS: next-generation sequencing; PZA: pyrazinamide; RIF: rifampicin; RRDR: rifampicin-resistance-determining region; SL-LPA: line probe assay for second-line drugs; TB: tuberculosis; WGS: wholegenome sequencing.
a Phenotypic DST should be performed for strains isolated from people during treatment monitoring. If resistance is detected, strains should be stored and WGS should be performed, if possible, to collect data on mutations associated with resistance.
b SL-LPAs do not cover the relevant region of the rrs gene or other genes associated with resistance to streptomycin.
Source: adapted or reproduced from Table 4 in Web Annex C.
 Feedback
Feedback