Book traversal links for A2.6 Information sheet: Practical considerations for implementation of the Nipro Genoscholar PZA-TB II assay
Nipro (Osaka, Japan) developed Genoscholar PZA-TB, a reverse hybridization-based technology for detection of pyrazinamide (PZA) resistance in tuberculosis (TB) (1, 2). Compared with MTBDRplus and MTBDRs/ LPA, the Genoscholar PZA-TB line-probe assay (LPA) does not include specific mutant probes, because resistance mutations are widespread across the entire pncA gene with no predominant mutations. Instead, the Genoscholar PZA-TB assay targets a 700 base pair (bp) fragment that covers the entire pncA gene and promoter region up to nucleotide -18 of the wild-type H37Rv reference strain that is known to harbour resistance-associated mutations. The first version of the assay contained 47 probes that covered the pncA promoter and open reading frame. The second version contained 48 probes. Three of the 48 probes (pncA 16, 17 and 35) in the second version represent silent mutations known to be genetic markers not associated with PZA resistance: Gly60Gly (probe 16), Ser65Ser (probe 17) and Thr142Thr (probe 35). The World Health Organization (WHO) includes this test as the first member within the class of high complexity reverse hybridization NAATs, and the recommendations below apply to this test (3).
WHO recommendations for use
In people with bacteriologically confirmed TB, high complexity reverse hybridization-based nucleic acid amplification tests (NAATs) may be used on culture isolates for detection of PZA resistance (rather than culture-based phenotypic drug susceptibility testing [DST]). (Conditional recommendation; very low certainty of evidence for diagnostic accuracy)
No special considerations are required in terms of subgroups (e.g. for children, people living with HIV [People with HIV] and those with extrapulmonary TB), given that the test is recommended for use on culture isolates.
Key performance conclusions
- The Nipro Genoscholar PZA-TB II test performs well for PZA resistance compared with phenotypic DST.
- The performance data on direct testing was limited and the recommendation currently only includes use on isolates.
- The pooled sensitivity and specificity data for the class are presented in a Web Appendix of the WHO consolidated guidelines (4).
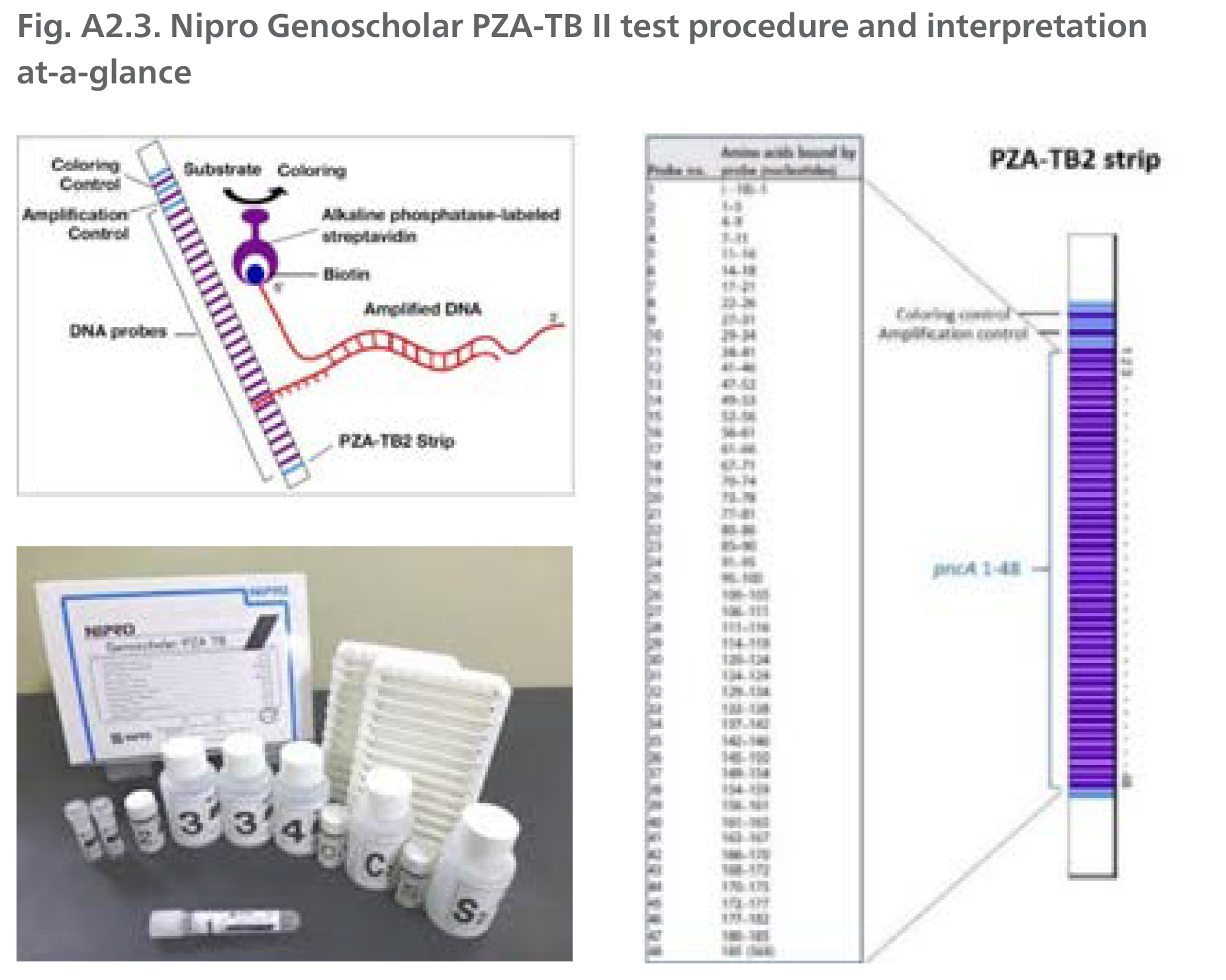
DNA is extracted from cultures, 5-10 µl of extract is amplified by polymerase chain reaction (PCR), denatured and hybridized using the MULTIBLOT NS-4800 system to complementary probes bound to a membrane-based strip. After hybridization, alkaline phosphatase-labelled streptavidin is added to bind any hybrids formed in the previous step and bound to the strip. The enzymatic reaction results in purple bands which are visually interpreted. The absence of wild type probe binding indicates the presence of a mutation.
Operational considerations
- Testing capacity: MULTIBLOTNS-4800 (48 strips per run), TwinCubator (12 strips per run)
- Storage temperature: 2-10 °C
- Shelf life: 18 months
- Unit price (consumable only): US$ 16 per test and US$ 14 000 per MULTIBLOTNS 4800 instrument
Implementation considerations
In addition to general guidance provided in Section 3.5, consider the following test-specific implementation considerations:
- Area 1 - Policies and planning: Genoscholar PZA-TB II integration into national algorithms and placement into networks should consider that the test should be placed at reference laboratories with adequate infrastructure; a well-functioning sample referral system from peripheral laboratories to reference laboratory should be in place; and patients with DR-TB (i.e. confirmed resistance to rifampicin [RIF] or isoniazid [INH]) may be prioritized for testing.
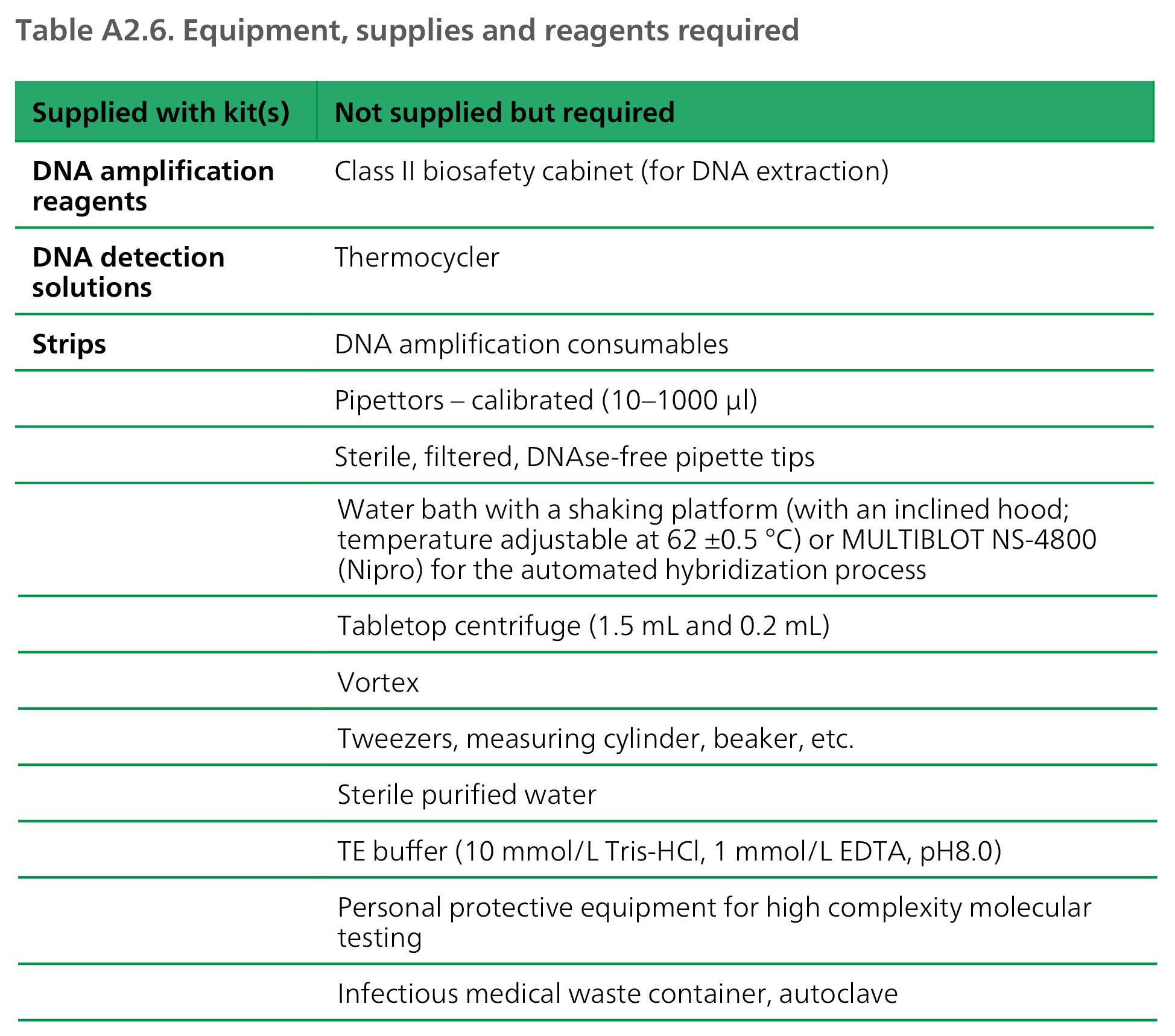
- Area 3 - Equipment: High complexity hybridization NAATs require multiple pieces of equipment for molecular processing (see Equipment, supplies and reagents above). High-throughput laboratories should consider procuring the Nipro automated MULTIBLOT NS-4800 instrument, which can increase testing capacity from 12 to 48 samples per run.
- Laboratory design and infrastructure: Precautions to reduce the risk of cross-contamination are critical. As a minimum requirement, three separate rooms for the different molecular steps should be established - one for DNA extraction, one for pre-amplification procedures, and one for amplification and post-amplification processes. Critical to attaining satisfactory results are restricted access, attention to the direction of workflow and meticulously followed procedures for cleaning.
- Area 5 - Procedures: Since mutations are only inferred by the absence of probes, the presence of mutations not associated with resistance may lead to reporting of resistance in the absence of resistance-associated mutations (false resistance). This limitation could be overcome by the sequencing of the pncA gene, especially if the pretest probability is low (e.g. RIF-susceptible TB case) and interpreting results based on the latest WHO catalogue of mutations.
- Area 7 - Quality assurance: High complexity reverse hybridization NAATs require strict adherence to a number of procedures to minimize the risk of contamination; therefore, the use of appropriate positive and negative controls, the monitoring of results based on expected outcomes to promptly detect false positive and false negative trends, and the regular participation to external quality assurance programmes should all be observed. New method validations should use PZA-sensitive and PZA-resistant, well-characterized isolates of Mycobacterium tuberculosis. PZA-resistant isolates should be selected to ensure a range of resistance-associated mutations are represented in the validation panel to ensure precision and accuracy of resistance detection can be achieved in relation to manufacturer-reported performance characteristics across differing resistance profiles.
- Area 9 - Training and competency assessment: Well-trained staff are needed to carry out a complex procedure that involves several manual steps, timed incubations, precise pipetting and care to avoid cross-contamination. In addition, special training and experience is required for reading of banding patterns on the strip and appropriate interpretation of results.
References for A2.6
- Sekiguchi J, Nakamura T, Miyoshi-Akiyama T, Kirikae F, Kobayashi I, Augustynowicz-Kopec E et al. Development and evaluation of a line probe assay for rapid identification of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis strains. J Clin Microbiol. 2007;45(9):2802-7 (https://pubmed.ncbi.nlm.nih.gov/17596354/).
- Nipro. Genoscholar PZA-TB II [website]. 2021 (https://www.nipro-group.com/en/products-services/genoscholartm-pza-tb-ii).
- Update on the use of nucleic acid amplification tests to detect TB and drug-resistant TB: rapid communication. Geneva: World Health Organization; 2021 (https://www.who.int/publications/i/item/update-on-the-use-of-nucleic-acid-amplification-tests-to-detect-tb-and-drug-resistant-tb-rapid-communication).
- WHO consolidated guidelines on tuberculosis Module 3: diagnosis - rapid diagnostics for tuberculosis detection. Geneva: World Health Organization; 2021 update (https://apps.who.int/iris/bitstream/handle/10665/342331/9789240029415-eng.pdf).
Source: Reproduced with permission of Nipro, © 2021. All rights reserved.
DNA: deoxyribonucleic acid; EDTA: ethylenediaminetetraacetic acid.
 Feedback
Feedback