Перекрёстные ссылки книги для Moderate complexity automated NAATs for detection of TB and resistance to rifampicin and isoniazid
Rapid detection of TB and rifampicin resistance is increasingly available as new technologies are developed and adopted by countries. However, what has also emerged is the relatively high burden of isoniazid-resistant, rifampicin-susceptible TB that is often undiagnosed. Globally, isoniazid-resistant, rifampicin-susceptible TB is estimated to occur in 13.1% (95% CI: 9.9–16.9%) of new cases and 17.4% (95% CI: 0.5–54.0%) of previously treated cases (14).
A new class of technologies has come to market with the potential to address this gap. Several manufacturers have developed moderate complexity automated NAATs for detection of TB and resistance to rifampicin and isoniazid on high throughput platforms for use in laboratories. The tests belonging to this class are faster and less complex to perform than phenotypic culturebased drug susceptibility testing (DST) and line probe assays (LPA). They have the advantage of being largely automated following the sample preparation step. Moderate complexity automated NAATs may be used as an initial test for detection of TB and resistance to both first-line TB drugs simultaneously (rifampicin and isoniazid). They offer the potential for the rapid provision of accurate results (important to patients) and for testing efficiency where high volumes of tests are required daily (important to programmes). Hence, these technologies are suited to areas with a high population density and rapid sample referral systems.
Recommendation
In people with signs and symptoms of pulmonary TB, moderate complexity automated NAATs may be used on respiratory samples for the detection of pulmonary TB, and of rifampicin and isoniazid resistance, rather than culture and phenotypic DST. Conditional recommendation, moderate certainty of evidence for diagnostic accuracy
There are several subgroups to be considered for this recommendation:
- The recommendation is based on evidence of diagnostic accuracy in respiratory samples of adults with signs and symptoms of pulmonary TB.
- The recommendation applies to people living with HIV (studies included a varying proportion of such individuals); performance on smear-negative samples was reviewed but was only available for TB detection, not for rifampicin and isoniazid resistance, and data stratified by HIV status were not available.
- The recommendation applies to adolescents and children based on the generalization of data from adults; an increased rate of indeterminate results may be found with paucibacillary TB disease in children.
- The review did not consider extrapolation of the finding for use in people with extrapulmonary TB and testing on non-sputum samples because data on diagnostic accuracy of technologies in the class for non-sputum samples were limited.
Test descriptions
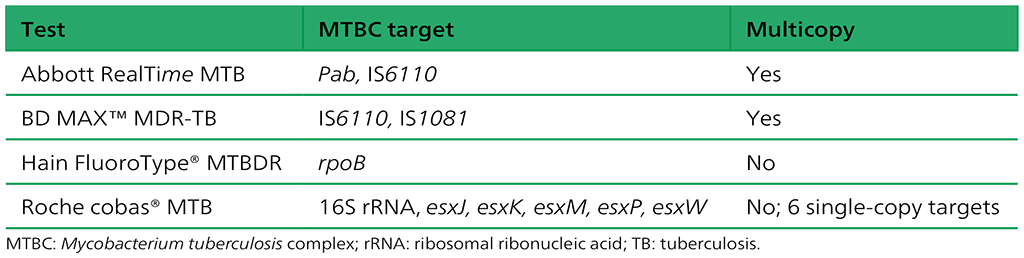
Abbott Molecular has two NAATs for TB, one for detection of Mycobacterium tuberculosis (Mtb) (RealTime MTB test), and one for detection of both rifampicin and isoniazid resistance (RealTime MTB RIF/INH). TB detection targets both the IS6110 genetic element and the pab gene. The rifampicin and isoniazid resistance test uses eight dye-labelled probes to detect variants in the rifampicin resistance determining region (RRDR) of the rpoB gene and four probes to detect isoniazid resistance, with two probes each for the katG and inhA genes. The company reports a limit of detection (LoD) of 17 cfu/mL for the RealTime MTB assay and of 60 colony forming units (cfu)/mL for the RealTime RIF/INH assay (14–16). The test is performed on the m2000 platform, m2000sp for automated DNA extraction and m2000rt for the real-time PCR.
Fig. 2.1.4. m2000sp RealTime system (a) and RealTime MTB Amplification Reagent Kit (b)
Becton Dickinson (BD) has a multiplexed real-time PCR (BD MAX™ MDR-TB) NAAT for the detection of Mtb and resistance to both rifampicin and isoniazid. The test is performed on a platform that uses five-colour detection (14). For Mtb detection, this test targets the multicopy genomic elements IS6110 and IS1081, as well as a single-copy genomic target. To detect resistance to rifampicin, the test targets the RRDR codons 507–533 Escherichia coli nomenclature (426–452 Mtb nomenclature) of the rpoB gene; for detection of resistance to isoniazid, the test targets both the inhA promoter region and the 315 codon of the katG gene. The LoD reported by the company is 0.5 cfu/mL for Mtb detection and 6 cfu/mL for resistance detection. The test is performed on the BD MAX platform, with the DNA automatically extracted and real-time PCR performed.
Fig. 2.1.5. BD MAX™ System (a) and BD MAX PCR Cartridges (b)
Bruker-Hain Diagnostics has two real-time NAATs: the FluoroType® MTB, which detects Mtb, and the FluoroType MTBDR, which detects Mtb and rifampicin and isoniazid resistance. These platforms are completely independent of the GenoType MTBDR platforms. The FluoroType MTBDR test uses asymmetric excess PCR and light on/off probes. The target genes are rpoB for detection of TB and rifampicin resistance and the inhA promoter and katG gene to detect isoniazid resistance. The LoD reported by the company is 15 cfu/mL for the FluoroType MTB test and 20 cfu/mL for the FluoroType MTBDR assay (14, 17, 18). For DNA extraction, manual (FluoroLyse) and automated (GenoXtract) options are available. The platforms used for amplification and detection are FluoroCycler® for the MTB assay and FluoroCycler XT for the MTBDR assay.
Fig 2.1.6. the FluoroType® MTB (a) and FluoroType MTBDR® (b) test principles
Roche Diagnostics (Roche) has two NAATs: cobas® MTB assay to detect Mtb, and cobas® MTB-RIF/INHassay to detect drug resistance (rifampicin and isoniazid) (14). The cobas MTB assay detects both 16S ribosomal RNA (rRNA) and esx genes as target genes for Mtb detection. The LoD reported by the company for this test was 7.6–8.8 cfu/mL. Rifampicin resistance is detected using RRDR and isoniazid resistance using the inhA promoter region and the katG gene. The tests are run on the cobas 6800/8800 systems, with the DNA automatically extracted and real-time PCR performed.
Figure 2.1.7 cobas® 6800 or 8800 system (a) and cobas® MTB Positive Control Kit (b)

Table 2.1.22. Mycobacterium genomic regions targeted by the different assays for TB detection included in the evaluation
In the moderate complexity class, an automated test is one that has (a) automated DNA extraction, (b) automated PCR preparation and (c) automated result interpretation, with either no pipetting steps or only one pipetting step between (a) and (c). These automated tests may require an initial manual specimen treatment step before the test material is transferred into the sample processing tube. Tests in the moderate complexity category require medical laboratories with biosafety measures in place and test-specific equipment; they also need welltrained, skilled and qualified laboratory staff to set up the tests and carry out the necessary equipment maintenance.
Justification and evidence
The WHO Global TB Programme initiated an update of the current guidelines and commissioned a systematic review on the use of moderate complexity automated NAATs for detection of TB and resistance to rifampicin and isoniazid in people with signs and symptoms of TB.
Three PICO questions were designed to form the basis for the evidence search, retrieval and analysis:
- Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of pulmonary TB, as compared with culture?
- Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of resistance to rifampicin, as compared with culture-based phenotypic DST?
- Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of resistance to isoniazid, as compared with culture-based phenotypic DST?
A comprehensive search of the following databases (PubMed, Embase, BIOSIS, Web of Science, LILACS and Cochrane) for relevant citations was performed. The search was restricted to the period January 2009 to July 2020. Reference lists from included studies were also searched. No language restriction was applied. Because there were few studies for the selected index tests, the diagnostic companies were contacted for reports of their internal validation data. Studies were also included from the WHO public call for submission of data. Mycobacterial culture was used as the reference standard for evaluation of Mtb detection. Resistance detection was compared with a phenotypic DST reference standard and a composite reference standard (that combines phenotypic and genotypic DST results) in studies where both had been performed.
Bivariate random-effects meta-analyses were performed using Stata software, to obtain pooled sensitivity and specificity estimates with 95% CIs for rifampicin resistance, isoniazid resistance and Mtb detection. Where only a limited number of studies were available, descriptive analyses were conducted.
For meta-analysis, studies were first meta-analysed separately for each test. Studies from all the tests were then used to obtain a pooled estimate for all technologies.
To decide whether pooling of all the tests would give meaningful estimates, various criteria for pooling were developed and agreed upon by the GDG panel before they were applied. Data were also evaluated and visualized using head-to-head comparisons of the tests with Xpert® MTB/RIF or any other WHO-recommended test.
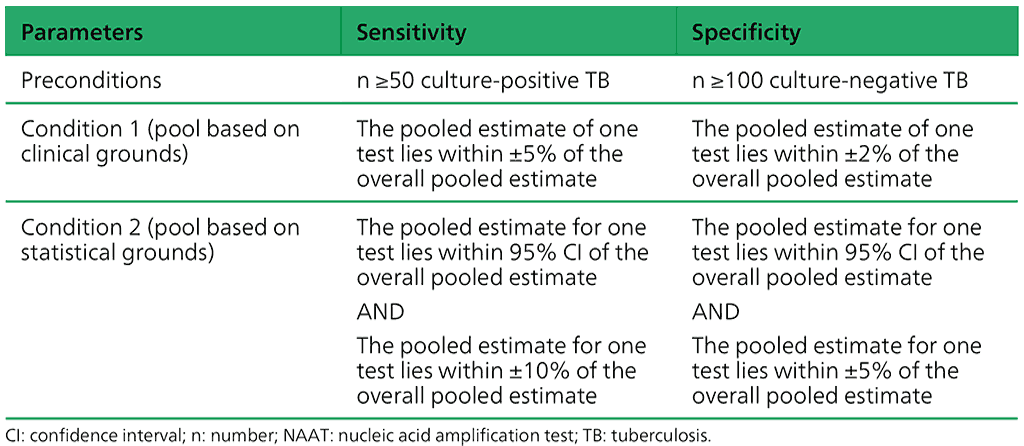
Data for all the index platforms were only pooled to answer PICO questions if they met the preconditions given in Table 2.1.23 and fulfilled either Condition 1 or Condition 2.
Table 2.1.23. Criteria for pooling studies on moderate complexity automated NAATs
The certainty of the evidence of the pooled studies was assessed systematically through PICO questions, using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) approach (19, 20). The GRADE approach produces an overall quality assessment (or certainty) of evidence and has a framework for translating evidence into recommendations; also, under this approach, even if diagnostic accuracy studies are of observational design, they start as high-quality evidence.
GRADEpro Guideline Development Tool software (19) was used to generate summary of findings tables. The quality of evidence was rated as high (not downgraded), moderate (downgraded one level), low (downgraded two levels) or very low (downgraded more than two levels), based on five factors: risk of bias, indirectness, inconsistency, imprecision and other considerations. The quality (certainty) of evidence was downgraded by one level when a serious issue was identified and by two levels when a very serious issue was identified in any of the factors used to judge the quality of evidence.
Data synthesis was structured around the three preset PICO questions, as outlined below. Three web annexes¹³ give additional information, as follows:
- details of studies included in the current analysis (Web Annex 1.3: Moderate complexity automated NAATs;
- a summary of the results and details of the evidence quality assessment (Web Annex 2.3: Moderate complexity automated NAATs); and
- a summary of the GDG panel judgements (Web Annex 3.3: Moderate complexity automated NAATs).
PICO 1: Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of pulmonary TB, as compared with culture?
A total of 29 studies with 13 852 specimens provided data for evaluating TB detection from the five index tests (Fig. 2.1.4). Of these 29 studies, 12 were conducted on the Abbott RealTime MTB test, six on FluoroType MTB, four on FluoroType MTBDR, five on BD MAX and two on the cobas MTB test. The reference standard for each of these studies for TB detection was mycobacterial culture.
Of the 29 studies, 16 (55%) had high or unclear risk of bias because they tested specimens before inclusion in the study, used convenience sampling or did not report the method of participant selection. Thus, the evidence was downgraded one level for risk of bias. Overall, the certainty of the evidence was moderate for sensitivity and high for specificity.
Fig. 2.1.8. Forest plot of included studies for TB detection with culture as the reference standard
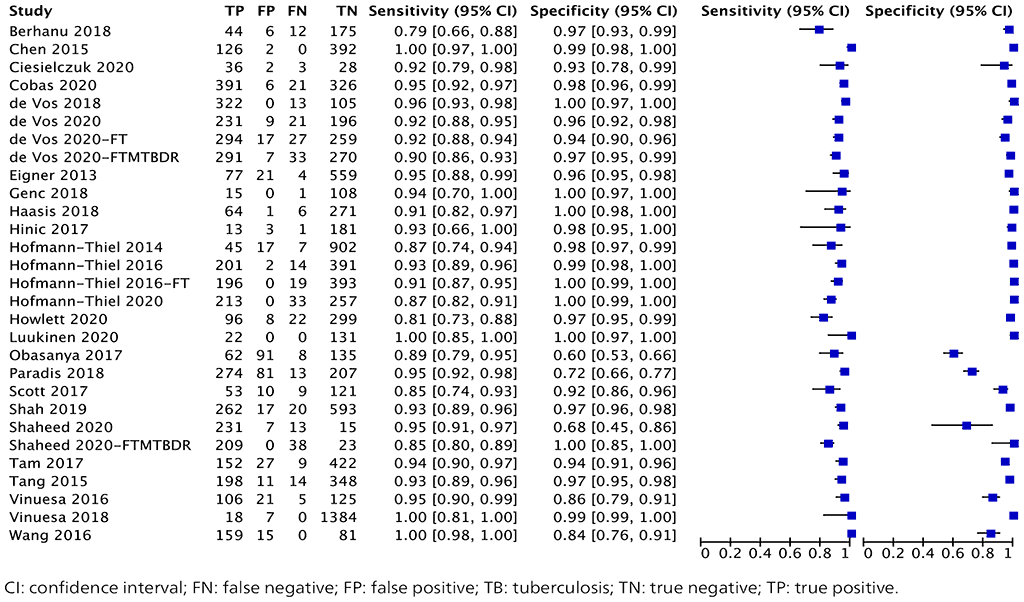
The overall sensitivity in these 29 studies ranged from 79% to 100%, and the specificity from 60% to 100%. The pooled sensitivity was 93.0% (95% CI: 90.9–94.7%) and the pooled specificity was 97.7% (95% CI: 95.6–98.8%).
PICO 2: Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of resistance to rifampicin, as compared with culture-based phenotypic DST?
A total of 18 studies with 2874 specimens provided data for resistance testing of rifampicin using moderate complexity automated NAATs (Fig. 2.1.5). Of these 18 studies, nine were conducted on the Abbott RealTime RIF/INH test, three on FluoroType MTBDR, four on BD MAX and two on the cobas RIF/INH test. The reference standard for each of these studies for resistance detection was phenotypic DST, using a composite reference standard with both phenotypic DST and sequencing results.
Eight (44%) of the 18 studies had high or unclear risk of bias because they did not report participant selection or tested specimens before inclusion in the study.
Fig. 2.1.9. Forest plot of included studies for rifampicin resistance detection with phenotypic DST as the reference standard
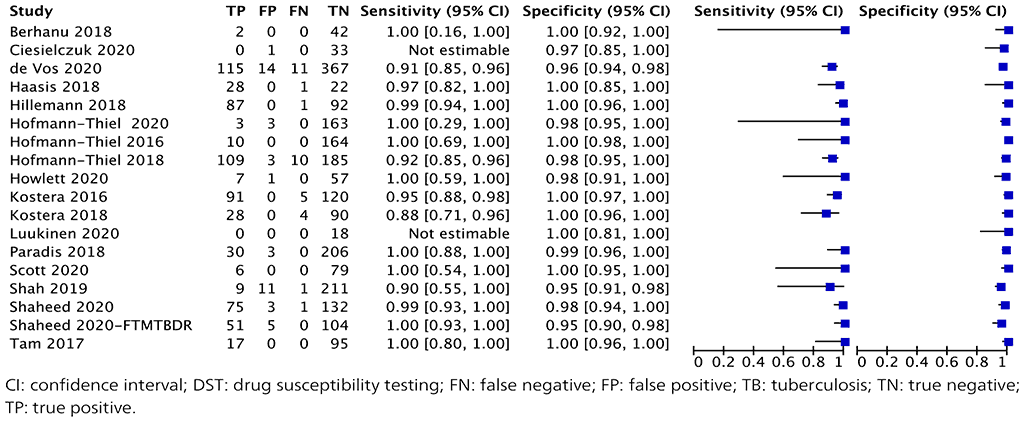
The overall sensitivity for rifampicin resistance in these 18 studies ranged from 88% to 100% and the specificity from 98% to 100%. The pooled sensitivity was 96.7% (95% CI: 93.1–98.4%) and the pooled specificity was 98.9% (95% CI: 97.5–99.5%).
In determining rifampicin resistance, the results from genetic sequencing (genotypic DST) were obtained where possible, and a composite reference standard was developed that combined the results from phenotypic and genotypic DST. For rifampicin resistance detection, the diagnostic test accuracy of moderate complexity automated NAATs was similar for phenotypic DST and the composite reference standard.
PICO 3: Should moderate complexity automated NAATs be used on respiratory samples in people with signs and symptoms of pulmonary TB for detection of resistance to isoniazid, as compared with culture-based phenotypic DST?
A total of 18 studies with 1758 specimens provided data for resistance testing of isoniazid using moderate complexity automated NAATs (Fig. 2.1.6). Of these 18 studies, nine were conducted on the Abbott RealTime RIF/INH test, three on FluoroType MTBDR, four on BD MAX and two on the cobas MTB-RIF/INH test. The reference standard for each of these studies for resistance detection was phenotypic DST, and a composite reference standard with both phenotypic DST and sequencing results.
Eight (44%) of the 18 studies had high or unclear risk of bias, because participant selection was not reported or prior testing was done on the included specimens.
Fig. 2.1.10. Forest plot of included studies for isoniazid resistance detection with phenotypic DST as the reference standard
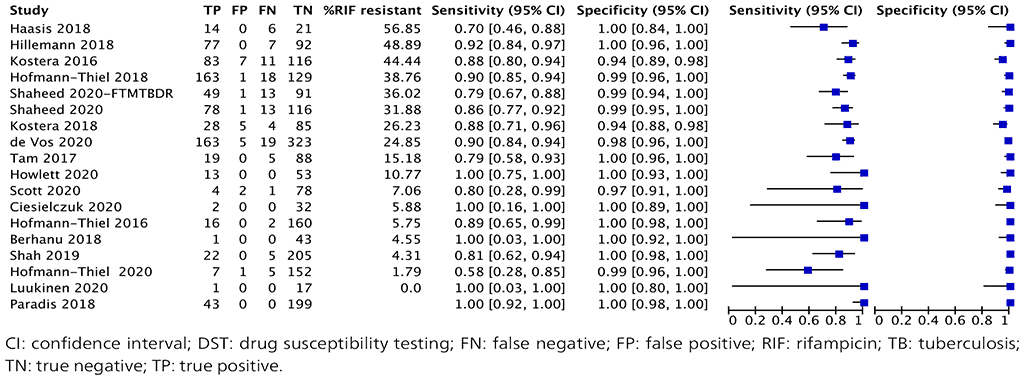
The overall sensitivity for isoniazid resistance in these 18 studies ranged from 58% to 100% and the specificity from 94% to 100%. The pooled sensitivity was 86.4% (95% CI: 82.1–89.8%) and the pooled specificity was 99.8% (95% CI: 98.3–99.8%).
In determining isoniazid resistance, the results from genetic sequencing (genotypic DST) were obtained where possible, and a composite reference standard was developed that combined the results from phenotypic and genotypic DST. For detecting isoniazid resistance, the diagnostic test accuracy of phenotypic DST was similar to that of the composite reference standard.
Cost-effectiveness analysis
This section answers the following additional question:
What is the comparative cost, affordability and cost–effectiveness of implementation of moderate complexity automated NAATs?
A systematic review was conducted, focusing on economic evaluations of moderate complexity automated NAATs. Four online databases (Embase, Medline, Web of Science and Scopus) were searched for new studies published from 1 January 2010 through 17 September 2020. The citations of all eligible articles, guidelines and reviews were reviewed for additional studies. Experts and test manufacturers were also contacted to identify any additional unpublished studies.
The objective of the review was to summarize current economic evidence and further understand the costs, cost–effectiveness and affordability of moderate complexity automated NAATs.
Several commercially available tests were included as eligible tests in the moderate complexity automated NAATs category; however, no published studies were identified assessing the costs or cost–effectiveness of any of those tests. One unpublished study comparing available data on two technologies from moderate complexity automated NAATs class was identified, and the data from that study are described below.
Unpublished data from FIND was provided through direct communication. This costing-only study used time and motion studies combined with a bottom-up, ingredients-based approach to estimate the unit test cost for the two selected technologies.¹⁴ Time and motion studies were conducted at a reference-level laboratory in South Africa. Several important simplifying assumptions were made that may limit the generalizability of the results; for example, 50% of laboratory operations dedicated to TB, a minimum daily throughput of 24 samples or the equivalent of one BD MAX run (24 tests/run), equipment costs fixed at US$ 100 000 for both platforms, a 5% annual maintenance cost, and the standard 3% discount rate and 10 years expected useful life years.
Additional literature searches conducted to look for economic data using similar platforms from non-TB disease areas identified three additional studies from HIV and hepatitis C virus (HCV) with limited cost data: one (20) using Abbott RealTime HIV and two on HCV (21, 22). Data were limited to cost per unit test kit and are not transferrable to test kit costs for the tests being considered in this review.
How large are the resource requirements (costs)?
Available unit test costs for two moderate complexity automated NAATs ranged from US$ 18.52 (US$ 13.79–40.70) and US$ 15.37 (US$ 9.61–37.40), with one study reporting cheaper per-test kit costs and higher operational costs associated with laboratory processing time. Equipment costs were strong drivers of cost variation and will vary across laboratory networks and operations. If equipment can be optimally placed or multiplexed to ensure high testing volume, the per-test cost can be minimized.
In one-way sensitivity analyses, annual testing volumes varied from fewer than 5000 tests/year to more than 25 000 tests/year. Per-test cost was highly sensitive to testing volume when fewer than 5000 tests were conducted per year; however, unit test costs begin to stabilize between 5000 and 10 000 tests/year, and above 10 000 tests/year, unit cost estimate was robust. When equipment can be multiplexed and used at capacity, per-test cost can be minimized.
What is the certainty of the evidence of resource requirements (costs)?
Available per-test cost data were unpublished but did include overheads, equipment, building, staff and consumable costs; however, complete quality assessment of the study was not possible. Test cost will vary according to testing volume and laboratory operations. There is limited evidence to assess the important variability across sites, countries and implementation approaches.
Does the cost-effectiveness of the intervention favour the intervention or the comparison?
No studies were identified that assessed cost–effectiveness for any of the moderate complexity automated NAATs, and extrapolation was not appropriate given differences in standard of care, care cascades and associated costs, operational conditions, testing volume and diagnostic accuracy. Implementation considerations (e.g. test placement, laboratory network and ability of the programme to initiate treatment quickly) are all likely to affect unit test cost and cost–effectiveness. Economic modelling is needed across various settings to understand the range of cost–effectiveness profiles of moderate complexity automated NAATs, and how they are likely to vary under different operational criteria.
Additional details on economic evidence synthesis and analysis are provided in Web Annex 4.9: Systematic literature review of economic evidence for NAATs to detect TB and DR-TB in adults and children.
User perspective
This section answers the following questions about key informants’ views and perspectives on the use of moderate complexity automated NAATs:
- Is there important uncertainty about or variability in how much end-users value the main outcomes?
- What would be the impact on health equity?
- Is the intervention acceptable to key stakeholders?
- Is the intervention feasible to implement?
User perspectives on the value, feasibility, usability and acceptability of diagnostic technologies are important in the implementation of such technologies. If the perspectives of laboratory personnel, clinicians, patients and TB programme personnel are not considered, the technologies risk being inaccessible to and underused by those for whom they are intended.
To address questions related to user perspective, two activities were undertaken:
- A systematic review of evidence on user perspectives and experiences with NAATs for detection of TB and TB drug resistance (moderate and low complexity automated assays, and high complexity hybridization-based assays) was undertaken from July to November 2020.
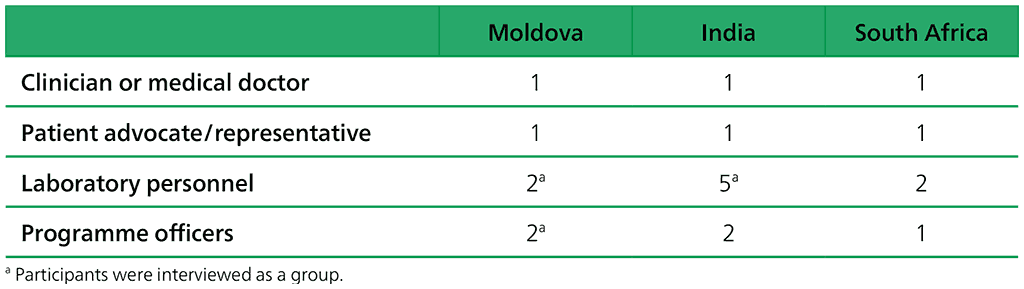
- A total of 14 semi-structured interviews with clinicians, programme officers, laboratory staff and patient advocates were conducted in India, Moldova and South Africa from October to November 2020.
The findings from these activities are discussed below.
Systematic review
A total of 27 studies were identified that met inclusion criteria, of which 21 were sampled for inclusion in the analysis. All of the sampled studies were published between 2012 and 2020. Of the 21 included studies, 18 were located in high TB burden countries: six in India, four in South Africa, two each in Kenya and Uganda, and one each in Brazil, Cambodia, Myanmar and Viet Nam. One study covered projects in nine countries (Bangladesh, Cambodia, Democratic Republic of the Congo, Kenya, Malawi, Moldova, Mozambique, Nepal and Pakistan). In addition, there was one study located in Eswatini, one in Mongolia and one in Nepal. All studies focused on Xpert MTB/RIF, except for one that focused on Xpert MTB/RIF Ultra (Xpert Ultra).
A summary of the core characteristics of studies included in this review is presented in a study characteristics table in Web Annex 4.10: User perspectives on NAATs to detect TB and DR-TB: results from qualitative evidence synthesis: systematic review.
Interviews
The aim of the interviews was to understand participants’ experiences of using the various technologies (i.e. NAATs for detection of TB and TB drug resistance) and their general TB diagnostic experiences. The three countries – India, Moldova and South Africa – were selected based on them being on WHO’s list of 30 high MDR-TB burden countries (2) and that index tests have been used to some extent in research contexts within these countries. Due to the short time frame, participants were purposively sampled and approached based on convenience through personal contacts and colleagues.
An overview of the participants is given in Table 2.1.24. To mask the identity of study participants they were coded by their country (Moldova [M], India [I] or South Africa [S]), their profession (clinician or medical doctor [M], patient advocate/representative [R], laboratory personnel [L] or programme officers [P]) and a number.
Table 2.1.24. Overview of participants for the end-users’ interviews
Interviews were conducted using Zoom, Skype or phone. Topics discussed included:
- current approach to diagnosing TB, MDR-TB and extensively drug-resistant TB (XDR-TB), including specific challenges;
- experiences with using molecular TB diagnostics and the index tests specifically, including details on steps taken in the diagnostic process;
- experiences with determining eligibility and treatment initiation, and challenges and benefits of using the index tests;
- overall usefulness of the index tests;
- the feasibility of implementing the index tests;
- the potential impact of the index tests on health equity; and
- how the potential impact of the index tests relates to current policy context.
Several important limitations of this approach were noted. Only a few participants were interviewed per country. Owing to the use of Zoom, Skype or phone for interviews, it was not possible to triangulate interview data with other evidence commonly collected through ethnographic approaches (e.g. multiple interviews and informal conversations at the same facility, observations or site visits). In addition, only some of the participants had personal experience with one or all of the index tests, and those participants who did have experience with the tests had used them in research settings rather than for routine practice.
More details on these interviews are given in Web Annex 4.10: User perspectives on nucleic acid amplification tests for tuberculosis and tuberculosis drug resistance: Interviews study.
Findings of the review and interviews
The main findings of the systematic review and interviews are given below. Where information is from the review, a level of confidence in the quality evidence synthesis (QES) is given; where it is from interviews, this is indicated with ‘Interviews’.
Is there important uncertainty about or variability in how much end-users value the main outcomes?
- Patients in high-burden TB settings value:
- getting an accurate diagnosis and reaching diagnostic closure (finally knowing "what is wrong with me");
- avoiding diagnostic delays because they exacerbate existing financial hardships and emotional and physical suffering, and make patients feel guilty for infecting others (especially children);
- having accessible facilities; and
- reducing diagnosis-associated costs (e.g. travel, missing work) as important outcomes of the diagnostic. QES: moderate confidenc
- Moderate complexity automated NAATs meet several preferences and values of clinicians and laboratory staff, in that they:
- are faster than culture-based phenotypic DST (similar to LPA or cartridge-based tests);
- have the advantage of being automated (unlike LPA);
- provide additional clinically relevant drug resistance information such as high versus low resistance (unlike the current Xpert MTB/RIF cartridge).
Interviews
What would be the impact on health equity?
- Various factors - for example, lengthy diagnostic delays, underuse of diagnostics, lack of TB diagnostic facilities at lower levels and too many eligibility restrictions - hamper access to prompt and accurate testing and treatment, particularly for vulnerable groups. QES: high confidence
- Staff and managers voiced concerns about:
- sustainability of funding and maintenance;
- complex conflicts of interest between donors and implementers; and
- the strategic and equitable use of resources, which negatively affects creating equitable access to cartridge-based diagnostics. QES: high confidence
- Access to clear and comprehensible information for TB patients on what TB diagnostics are available to them and how to interpret results is a vital component of equity, and lack of such access represents an important barrier for patients.
Interviews - New treatment options need to be matched with new diagnostics. It is important to improve access to treatment based on new diagnostics and to improve access to diagnostics for new treatment options.
Interviews - The speed at which WHO guidelines are changing does not match the speed at which many country programmes are able to implement the guidelines. This translates into differential access to new TB diagnostics and treatment:
- between countries (i.e. between those that can and cannot quickly keep up with the rapidly changing TB diagnostic environment); and
- within countries (i.e. between patients who can and cannot afford the private health system that is better equipped to quickly adopt new diagnostics and policies). Interviews
- The identified challenges with the use of NAATs for detection of TB and DR-TB, and accumulated delays, risk compromising the added value as identified by the users, ultimately leading to underuse. The challenges also hamper access to prompt and accurate testing and treatment, particularly for vulnerable groups.
QES: high confidence
Is the intervention acceptable to key stakeholders?
- Patients can be reluctant to test for TB or MDR-TB because of:
- stigma related to MDR-TB or having interrupted treatment in the past;
- fears of side-effects;
- failure to recognize symptoms;
- inability to produce sputum; and
- cost, distance and travel concerns related to (repeat) clinic visits.
QES: high confidence
- Health workers can be reluctant to test for TB or MDR-TB because of:
- TB-associated stigma and consequences for their patients;
- fear of acquiring TB;
- fear from supervisors when reclassifying patients already on TB treatment who turn out to be misclassified;
- fear of side-effects of drugs in children; and
- community awareness of disease manifestations in children.
QES: high confidence
- In relation to the acceptability of moderate complexity automated NAATs:
- the automation of this class of technologies, which recognizes the high workload of laboratory staff, improves their acceptability;
- in terms of the physical size of the platform and how it fits into the laboratory space and workflow, a smaller footprint may be more acceptable; and
- the number of samples run on the system is acceptable provided that the platform is placed within a laboratory that receives a sufficient sample load to run the system.
Interviews
Is the intervention feasible to implement?
- The feasibility of all diagnostic technologies is challenged if there is an accumulation of diagnostic delays or underuse (or both) at every step in the process, mainly because of health system factors such as:
- non-adherence to testing algorithms, testing for TB or MDR-TB late in the process, empirical treatment, false negatives due to technology failure, large sample volumes and staff shortages, poor or delayed sample transport and sample quality, poor or delayed communication of results, delays in scheduling follow-up visits and recalling patients, and inconsistent recording of results;
- lack of sufficient resources and maintenance (i.e. stock-outs; unreliable logistics; lack of funding, electricity, space, air conditioners and sputum containers; dusty environment; and delayed or absent local repair option);
- inefficient or unclear workflows and patient flows (e.g. inefficient organizational processes, poor links between providers, and unclear follow-up mechanisms or information on where patients need to go); and
- lack of data-driven and inclusive national implementation processes.
QES: high confidence
- The feasibility of moderate complexity automated NAATs is also challenged by:
- how or whether the platform fits into the physical space of the laboratory (considering bench size and weight of the platform) and sample workflow;
- a poorly functioning sample transport system that affects the quality of samples; and
- the need to ensure that clinicians and laboratory staff have time to communicate effectively regarding diagnostic results if the platform is centralized, while also ensuring that the laboratory location is central enough to receive adequate numbers of samples to make the machine worth running.
Interviews
- Implementation of new diagnostics must be accompanied by training for clinicians to help them interpret results from new molecular tests and understand how this information is translated into prompt and proper patient management. In the past, with the introduction of Xpert MTB/RIF, this has been a challenge.
QES: high confidence and interviews - Introduction of new diagnostics must be accompanied by guidelines and algorithms that support clinicians and laboratories in communicating with each other, such that they can discuss discordant results and interpret laboratory results in the context of drug availability, patient history and patient progress on a current drug regimen.
Interviews
Implementation considerations
Factors to consider when implementing moderate complexity automated NAATs for detection of TB and resistance to rifampicin and isoniazid are as follows:
- local epidemiological data on resistance prevalence should guide local testing algorithms, whereas pretest probability is important for the clinical interpretation of test results;
- the cost of a test varies depending on parameters such as the number of samples in a batch and the staff time required; therefore, a local costing exercise should be performed;
- low, moderate and high complexity tests have successive increase in technical competency needs (qualifications and skills) and staff time, which affects planning and budgeting;
- availability and timeliness of local support services and maintenance should be considered when selecting a provider;
- laboratory accreditation and compliance with a robust quality management system (including appropriate quality control) are essential for sustained service excellence and trust;
- training of both laboratory and clinical staff is needed to ensure effective delivery of services and clinical impact;
- use of connectivity solutions for communication of results is encouraged, to improve efficiency of service delivery and reduce time to treatment initiation;
- moderate complexity automated NAATs may already be used programmatically for other diseases - for example, severe acute respiratory syndrome coronavirus 2 (SARS-CoV2), HIV and antimicrobial resistance (AMR) - which could potentially facilitate implementation of TB testing on shared platforms;
- implementation of moderate complexity automated NAATs requires laboratories with the required infrastructure, space and efficient sample referral systems;
- although these are automated tests, well-trained skilled staff are needed to set up assays and complete maintenance requirements; and
- implementation of these tests should be context specific; thus, it should take into account access issues, especially in remote areas, where less centralized WHO-recommended technologies may be more appropriate.
Research priorities
Research priorities for moderate complexity automated NAATs for detection of TB and resistance to rifampicin and isoniazid are as follows:
- diagnostic accuracy in specific patient populations (e.g. children, people living with HIV, and patients with signs and symptoms of extrapulmonary TB) and in non-sputum samples;
- impact of diagnostic technologies on clinical decision-making and outcomes that are important to patients (e.g. cure, mortality, time to diagnosis and time to start treatment) in all patient populations;
- impact of specific mutations on treatment outcomes among people with DR-TB;
- use, integration and optimization of diagnostic technologies in the overall landscape of testing and care, as well as diagnostic pathways and algorithms;
- economic studies evaluating the costs, cost-effectiveness and cost-benefit of different diagnostic technologies;
- qualitative studies evaluating equity, acceptability, feasibility and end-user values of different diagnostic technologies;
- effect of non-actionable results (indeterminate, non-determinate or invalid) on diagnostic accuracy and outcomes that are important to patients;
- operational research on the advantages and disadvantages of individual technologies within the class of moderate complexity automated NAATs;
- effect of moderate complexity automated NAATs in fostering collaboration and integration between disease programmes; and
- the potential utility of detecting katG resistance to identify MDR-TB clones that may be missed because they do not have an RRDR mutation (e.g. the Eswatini MDR-TB clone, which has both the katG S315T and the non-RRDR rpoB I491F mutation).
13 A complete list of web annexes is provided at page iv–vi.
14 Data courtesy of H Sohn and W Stevens at FIND (unpublished).
 Обратная связь
Обратная связь