Liens transversaux de livre pour 1.1 Background
The political declaration at the first United Nations (UN) high-level meeting on tuberculosis (TB) held on 26 September 2018 included commitments by Member States to four new global targets (1), which were subsequently renewed at the second UN high-level meeting on TB on 22 September 2023 (2). One of these targets is that at least 90 per cent of the estimated number of people who develop TB are reached with quality-assured diagnosis and treatment in the 5-year period 2023–2027 (2).
In 2022, 7.5 million people were diagnosed with TB (3). Globally, in the same year, the estimated number of people who developed MDR-TB or RR-TB (MDR/RR-TB) was 410 000 (95% UI: 370 000–450 000).
The WHO’s End TB Strategy calls for the early diagnosis of TB and universal drug susceptibility testing (DST), highlighting the critical role of laboratories in the post-2015 era in rapidly and accurately detecting TB and drug resistance (4).
WHO has endorsed a range of new diagnostic technologies during the past 16 years (Table 1.1). The WHO assessment process for TB diagnostics currently focuses on evaluating classes of TB diagnostic technologies rather than specific products.
Table 1.1. Classes of technologies and associated products included in current guidelines
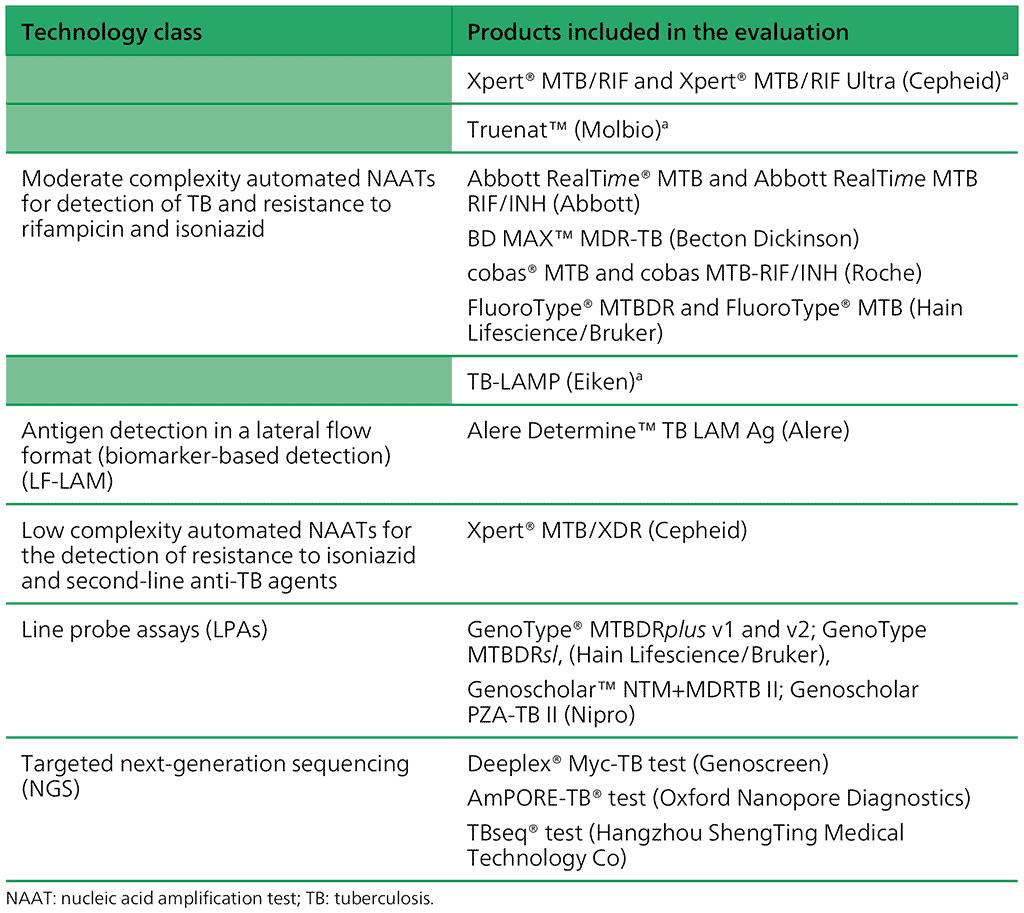
a These recommendations are currently product specific but will be changed to be class based, to align with the other recommendations.
Real-time polymerase chain reactions (PCR) methods. These technologies include tools such as Xpert® MTB/RIF and Xpert® MTB/RIF Ultra (Cepheid) or Truenat™ (Molbio) that are automated and provide an all-in-one solution suitable for the peripheral level are the most widely used today. These tools detect MTBC DNA and can detect mutations in the gene associated with rifampicin resistance. The available tools use software and hardware (computers) to report results and require well-established laboratory networks and trained personnel.
Moderate complexity automated nucleic acid amplification methods (NAATs) for detection of TB and resistance to rifampicin and isoniazid. The tests belonging to this class are faster and less complex to perform than phenotypic culture-based drug susceptibility testing (DST) and line probe assays (LPA). They have the advantage of being largely automated following the sample preparation step. Moderate complexity automated NAATs may be used as an initial test for detection of TB and resistance to rifampicin and isoniazid simultaneously. They offer the potential for the rapid provision of accurate results and for testing efficiency where high volumes of tests are required daily. Hence, these technologies are suited to areas with a high population density and rapid sample referral systems.
Loop-mediated isothermal amplification (TB-LAMP). This technology is based on DNA amplification at a single temperature range (in contrast to the PCR, which requires a thermocycler). Detection of amplified product is done visually, using an ultraviolet (UV) lamp, directly in the reaction tubes. The method requires only basic equipment and can be implemented at the lowest levels of the laboratory network. However, detection of mutations in resistance-associated genes is not available with the currently recommended technology
Antigen detection in a lateral flow format (biomarker-based detection) (LF-LAM). The currently available urinary LAM assays have suboptimal sensitivity and specificity and are therefore not suitable as diagnostic tests for TB in all populations. However, unlike traditional diagnostic methods, urinary LAM assays demonstrate improved sensitivity for the diagnosis of TB among individuals coinfected with HIV.
Low complexity automated NAATs for the detection of resistance to isoniazid and second-line anti-TB agents. This is a new class of diagnostics intended for use as a reflex test in specimens determined to be Mtb complex (MTBC)-positive; they offer rapid DST in intermediate and peripheral laboratories. The first product in this class simultaneously detects resistance to isoniazid, fluoroquinolones, ethionamide and amikacin. Results are available in under 90 minutes, leading to faster time to results than the current standard of care, which includes LPAs and culture-based phenotypic DST.
Line probe assays (LPAs). These are a family of DNA strip-based tests that can detect the MTBC DNA and determine its drug-resistance profile. The tests do this through the pattern of binding of amplicons (DNA amplification products) to probes that target the specific parts of the MTBC genome, common resistance-associated mutations to anti-TB drugs or the corresponding wildtype DNA sequence (5). LPAs are technically more complex to perform than the Xpert MTB/ RIF assay; however, they can detect resistance to a broader range of first-line and second-line agents and provide mutation specific data for common variants. Testing platforms have been designed for a reference laboratory setting and are most applicable to high TB burden countries. Results can be obtained in 5 hours (5).
Targeted next-generation sequencing (NGS). The present update includes a new class of tests – targeted next-generation sequencing (NGS) – which can be used for the detection of drug resistance to the broader list of drugs. This class of tests is based on technology that couples amplification of selected genes with NGS to detect resistance to many drugs with a single test. Also, since targeted NGS can interrogate entire genes to identify specific mutations associated with resistance, the accuracy may be better than that of existing WHO-recommended diagnostic tests (WRDs). In addition, new tests based on targeted NGS can detect resistance to new and repurposed drugs not currently included in any other molecular assays. Hence, this class of tests offers great potential to provide comprehensive resistance detection matched to modern treatment regimens.
Regulatory approval from national regulatory authorities or other relevant bodies is required before implementation of new diagnostic tests.
 Retour
Retour