Liens transversaux de livre pour 4.4.1 Decision pathway for Algorithm 4
- WHO guidelines stress the importance of DST before treatment, especially for medicines for which mWRDs are available.
- People with TB that is RIF susceptible, INH susceptible or unknown should be started on a first-line regimen for drug-susceptible TB (55).
- Globally, Hr-TB prevalence is 7.4% (95% CI: 6.5–8.4%) in new cases and 11.4% (95% CI: 9.4–13.4%) in people who were treated previously (56). The prevalence in some settings can exceed 25%. Contacts of a person known to have Hr-TB are also at increased risk. The prevalence of any INH resistance is particularly high in some parts of the WHO European Region and Western Pacific Region.
- Hr-TB is currently undetected in many settings but is clinically important. Compared with people with drug-susceptible TB, people with Hr-TB who are treated with the recommended regimen for drug-susceptible TB have a much higher risk of treatment failure (11% versus 2%), relapse (10% versus 5%) and acquiring additional drug resistance (8% versus 1%) (56).
- The successful treatment of Hr-TB, prevention of the spread of Hr-TB and acquisition of resistance to additional drugs such as RIF rely on rapidly detecting people with Hr-TB and placing them on effective treatment regimens. The low complexity automated NAATs for follow-up detection of INH resistance can be valuable tools owing to their ease of use and the possibility of implementing these tests in the lower levels of the health system.
- The recommended Hr-TB treatment regimen is RIF, EMB, PZA and LFX for 6 months (9, 57).
- Targeted NGS tests report results for many medicines not used for treatment of drug-susceptible TB (e.g. BDQ, LZD, CFZ, AMK and STR). These results should not be released for people with RIF-susceptible TB; however, where this is not possible, the results should make clear that these medicines are only to be used for individualized regimens in specialized circumstances.
- Reliable phenotypic DST methods are available for RIF, INH, FQs, BDQ, CFZ, Pa, CS, LZD, AMK and DLM. Testing algorithms that rely on culture and phenotypic DST are described in the WHO policy framework (53) and technical manual (Web Annex C). Member States should ensure there is capacity for DST for drugs used for treatment and for which reliable testing is available.
- No reliable phenotypic DST methods are available for EMB, ETO/prothionamide, or imipenem-cilastatin/meropenem; hence, results should not be used for clinical decision-making.
- Initiation of treatment should not be delayed while waiting for the results of DST.
Fig. 4.7. New Algorithm 4: Follow-on testing for people with RIF-susceptible TB at risk of resistance to other drug
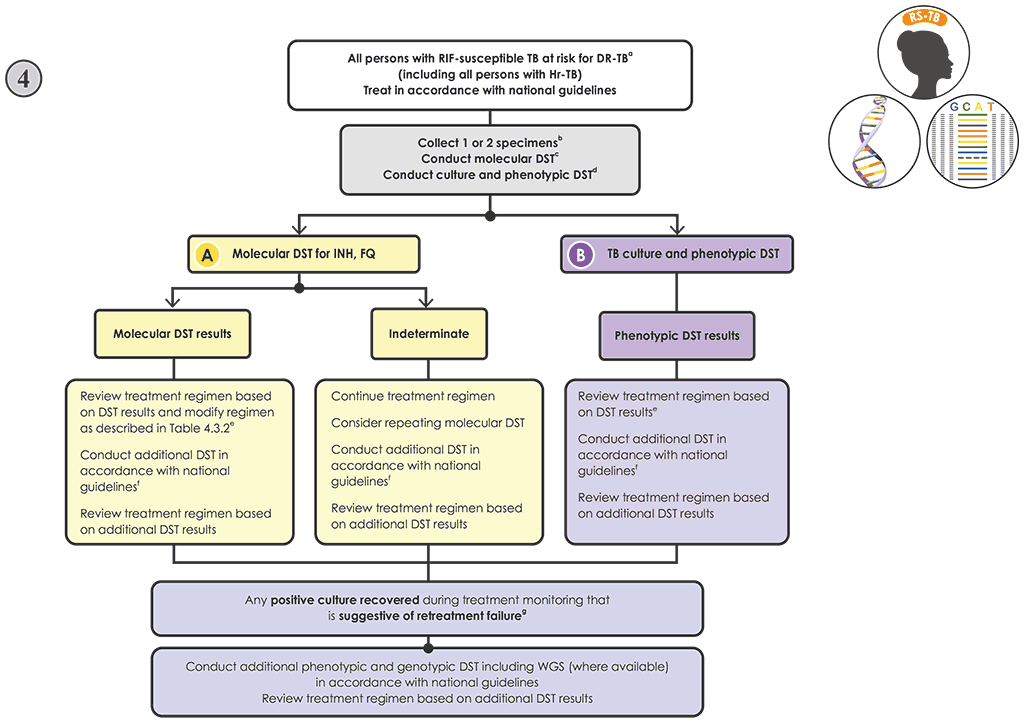
Decision pathway for Algorithm 4 – Follow-on testing for individuals with RIF-susceptible TB at risk of resistance to other drugs
- Promptly initiate the person on a regimen for the treatment of RIF-susceptible TB in accordance with national guidelines (55). Individuals with Hr-TB should be started on an Hr-TB regimen (9, 57).
- If molecular and phenotypic testing are performed in the same laboratory, collecting one specimen may be sufficient. If testing is performed in two laboratories, collect two specimens and conduct the molecular and phenotypic testing in parallel. Transport sputum specimens or isolates to the appropriate testing laboratory, if necessary.
- Conduct molecular testing, and perform culture in parallel.
- If targeted NGS tests are used:
- Modify the treatment if appropriate, and perform phenotypic DST when the culture is positive based on Table 4.3.2. Note: the results from sequencing produce information on multiple drugs simultaneously; however, for simplicity, Table 4.3.2 takes a single-drug approach for interpreting targeted NGS test results, although all results should be taken into consideration when designing a treatment regimen.
- If targeted NGS tests are used:
Table 4.3.2 Treatment modifications and follow-on DST for Hr-TB based on results from targeted NGS
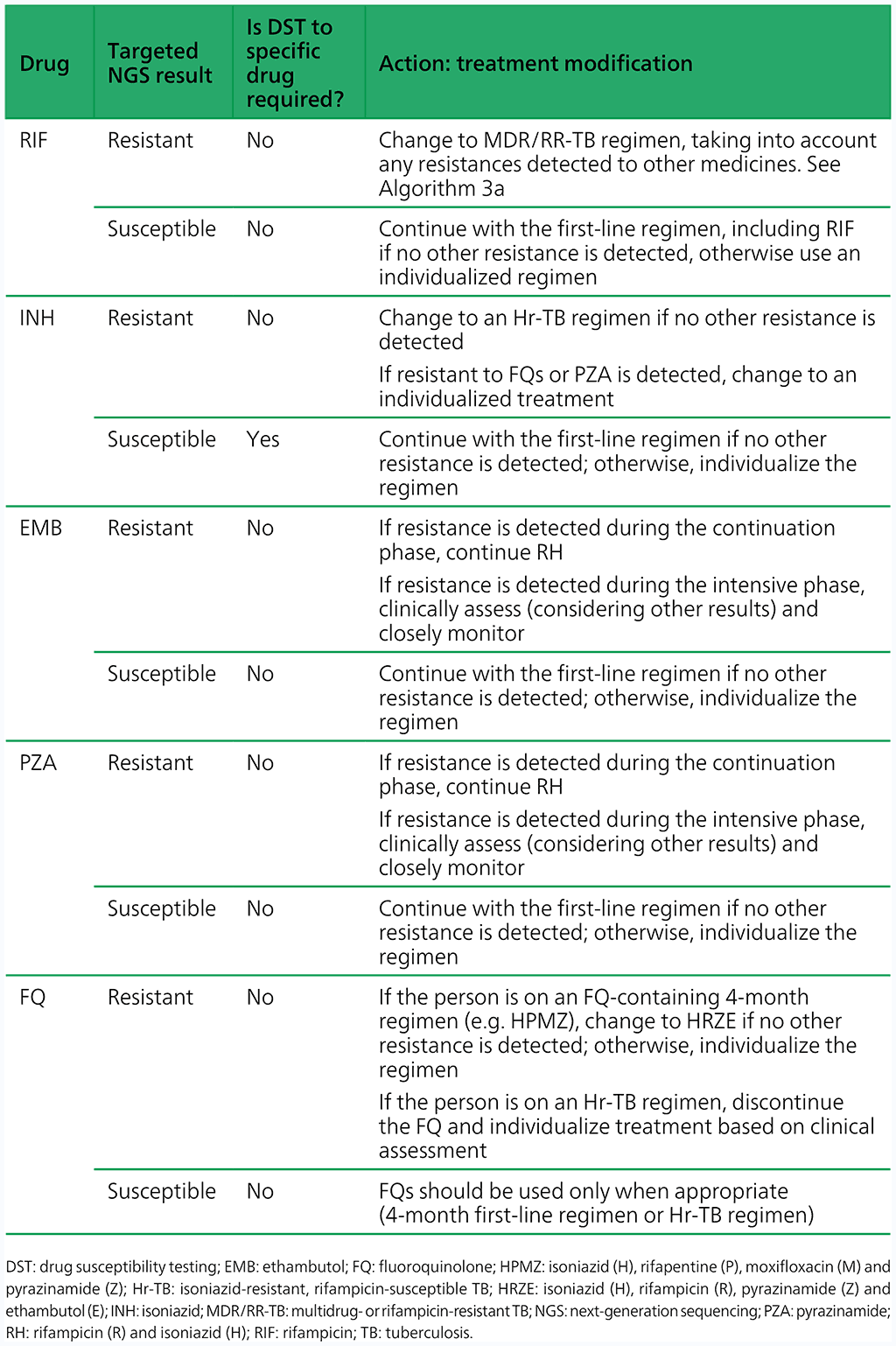
- If the targeted NGS test result is indeterminate, the targeted NGS test should be repeated with a fresh sample, and treatment decisions based on clinical assessments, epidemiologic situation and the results of phenotypic DST.
- If a WHO recommended follow-on molecular test other than targeted NGS tests is used:
- For a person with RR-TB by DST, detected by either a molecular (e.g. Xpert MTB/RIF, Xpert Ultra or Truenat) or phenotypic DST, but no results are available for INH and the person is at high risk for Hr-TB, start at Step 1 below.
- For a person who had an initial TB test that included RIF and INH results (e.g. a moderate complexity automated NAAT was used) in Algorithm 1, go to Step 4 below.
- Collect a good-quality specimen and transport it to the testing laboratory for molecular or phenotypic testing for INH resistance:
- Testing could follow a two-step process: detection of INH resistance followed by detection of FQ resistance. The two-step process is applicable when a moderate complexity automated NAAT or FL-LPA is used for Hr-TB detection, followed by the low complexity automated NAAT or SL-LPA for FQ resistance detection. A single step option is now available using the first-in-class low complexity automated NAAT, which detects both INH and FQ resistance simultaneously.
- Phenotypic DST may be required for INH resistance determination because of the sensitivity; depending on the test used, it may miss about 15% of resistant samples (Table 3.3 in Section 3). Phenotypic DST will be relevant when the person is at high risk for Hr-TB. If both molecular and phenotypic tests are performed, initiate the tests in parallel; do not wait for the results of one test before initiating the other test.
- Culture-based phenotypic DST for INH requires 3–8 weeks to produce a result. Phenotypic DST may be useful for evaluating people with results from an mWRD showing susceptibility to INH, particularly in populations with a high pretest probability for resistance to INH.
- If INH resistance is not detected, continue treatment with a first-line regimen in accordance with national guidelines:
- Conduct additional DST in accordance with national guidelines.
- Consider requesting additional molecular or phenotypic DST for resistance to INH if the person is thought to be at risk of having Hr-TB, despite the mWRD result.
- If INH resistance is detected:
- Using the low complexity automated NAAT will provide simultaneous detection of resistance to INH and FQ. If FQ resistance is not detected, follow Steps 3b and 5. If FQ resistance is detected, follow Steps 3d(ii) and 5. If the FQ result is unknown or unsuccessful, follow Step 3c.
- Initiate treatment with an Hr-TB regimen (9):
- There is no clear evidence showing that adding INH at the usual doses adds benefits or harms to people. For the convenience of people being treated and for ease of administration, the four-drug INH/RIF/EMB/PZA (HREZ) fixed-dose combination tablets may be used to deliver the Hr-TB regimen, alongside LFX.
- According to emerging evidence, people infected with strains with only inhA promoter mutations and corresponding modest increases in minimal inhibitory concentration (MIC) may benefit from high-dose INH therapy. Thus, additional INH – up to a maximum dose of 15 mg/kg per day – may be considered for use with the Hr-TB regimen for such isolates. The added value of isoniazid in the regimen, even when used at the higher dose, declines as MICs increase further.
- Refer a specimen from a person with laboratory-confirmed Hr-TB for molecular (e.g. low complexity automated NAAT or SL-LPA) or phenotypic DST for FQs and PZA. Note: if the Xpert MTB/XDR test was used in Step 1, the FQ result will already be available, so go to Step 3d.
- Rapid molecular testing for FQ resistance is preferred. When used for direct testing of sputum specimens, the low complexity automated NAAT and SL-LPA detects 93% and 86% of people with FQ resistance, respectively (Table 3.4 in Section 3):
- Low complexity automated NAATs provide rapid results and are suitable for use at the peripheral level. The first-in-class test, Xpert MTB/XDR, reports low-level FQ resistance when the mutations gyrA A90V, gyrA S91P and gyrA D94A are detected from the probe melting temperature (49). Phenotypic DST at the clinical breakpoint for MFX should be performed to confirm the potential value of high-dose MFX treatment for such people.
- The diagnostic accuracy of SL-LPA is similar when it is performed directly on sputum or cultured isolates. SL-LPA can be used with smear-positive or smear-negative specimens, although a higher indeterminate rate will occur when testing smear-negative specimens.
- Despite good specificity and sensitivity of low complexity automated NAATs and SL-LPA for the detection of FQ resistance, phenotypic DST is required to exclude resistance to individual FQs completely. In particular, phenotypic DST may be needed in settings with a high pretest probability for resistance to FQ, to exclude resistance when the SL-LPA does not detect mutations associated with resistance.
- Review FQ resistance results:
- If FQ resistance is not detected, continue treatment with the LFX-containing Hr-TB regimen.
- If FQ resistance is detected:
- Discontinue use of LFX and change to a 6-month regimen of (INH)/RIF/EMB/ PZA; that is, 6(H)REZ, where the “(H)” indicates that the INH is optional) or an individualized Hr-TB regimen.
- Refer a specimen for PZA DST if reliable PZA DST is available in the country. Options include the high complexity reverse hybridization NAAT, phenotypic DST in the MGIT system and pncA sequencing. For more details, see the WHO technical manual for drug susceptibility testing of medicines used in TB treatment (Web Annex C).
- If PZA resistance is not detected, or if PZA DST is not available, continue therapy with the regimen that was designed based on the previous DST results.
- If PZA resistance is detected, it may be necessary to design individualized treatment regimens, especially if resistance to both FQs and PZA is detected.
- Rapid molecular testing for FQ resistance is preferred. When used for direct testing of sputum specimens, the low complexity automated NAAT and SL-LPA detects 93% and 86% of people with FQ resistance, respectively (Table 3.4 in Section 3):
- If the INH result cannot be interpreted or is invalid, repeat the low or moderate complexity automated NAAT or FL-LPA with a fresh specimen. Consider conducting culture and molecular or phenotypic DST for INH on the isolate, if the person is considered to be at risk of having Hr-TB.
- For all people with TB, treatment monitoring should include collecting samples for culturing, as described in WHO guidelines. Any positive culture suggestive of treatment failure should undergo phenotypic and molecular DST, if available. At a minimum, DST should include testing for resistance to INH and RIF for people on first-line regimens, and for RIF, FQs and PZA (if available) for people on Hr-TB regimens. The treatment regimen should be modified as necessary, based on the results of the DST.
- Collect a good-quality specimen and transport it to the testing laboratory for molecular or phenotypic testing for INH resistance:
 Retour
Retour