Liens transversaux de livre pour High complexity reverse hybridization-based NAATs for detection of pyrazinamide resistance
Pyrazinamide is an important antibiotic for the treatment of both drug-susceptible TB and DR-TB because of its unique ability to eradicate persisting bacilli and its synergistic properties with other antibiotics. Mono-resistance to pyrazinamide is rare; however, pyrazinamide resistance is strongly associated with MDR/RR-TB, with an estimated 30–60% of MDR/RR-TB also resistant to pyrazinamide. Thus, for people diagnosed with RR-TB, it is important to detect the presence of pyrazinamide resistance so that clinicians can make an informed decision on whether to include or exclude pyrazinamide in the treatment regimen. The high complexity hybridization-based NAAT may be used for diagnosis of pyrazinamide resistance on patient isolates; however, performance of this test requires appropriate infrastructure and skilled staff.
Recommendation
In people with bacteriologically confirmed TB, high complexity reverse hybridization-based NAATs may be used on Mtb culture isolates for detection of pyrazinamide resistance rather than culture-based phenotypic DST.
Conditional recommendation, very low certainty of evidence for diagnostic accuracy
In terms of subgroups to be considered for this recommendation, no special considerations are required (e.g. for children, people living with HIV and those with extrapulmonary TB), given that the test is recommended for use on culture isolates.
Test description
Nipro (Osaka, Japan) developed Genoscholar™ PZA-TB, an LPA with reverse hybridization-based technology for detection of pyrazinamide resistance (43). This assay is a commercially available rapid molecular test for detection of pyrazinamide resistance. Compared with MTBDRplus and MTBDRsl LPA, the Genoscholar PZA-TB LPA does not include specific mutant probes because resistance mutations are widespread across the entire pncA gene with no predominant mutations. Instead, the Genoscholar PZA-TB assay targets a 700 base pair (bp) fragment covering the entire pncA gene and promoter region up to nucleotide –18 of the wild-type H37Rv reference strain.
Fig. 2.3.7. Nipro GenoScholar PZA-TB II strip (a) and Nipro GenoScholar PZA-TB II kit contents (b)
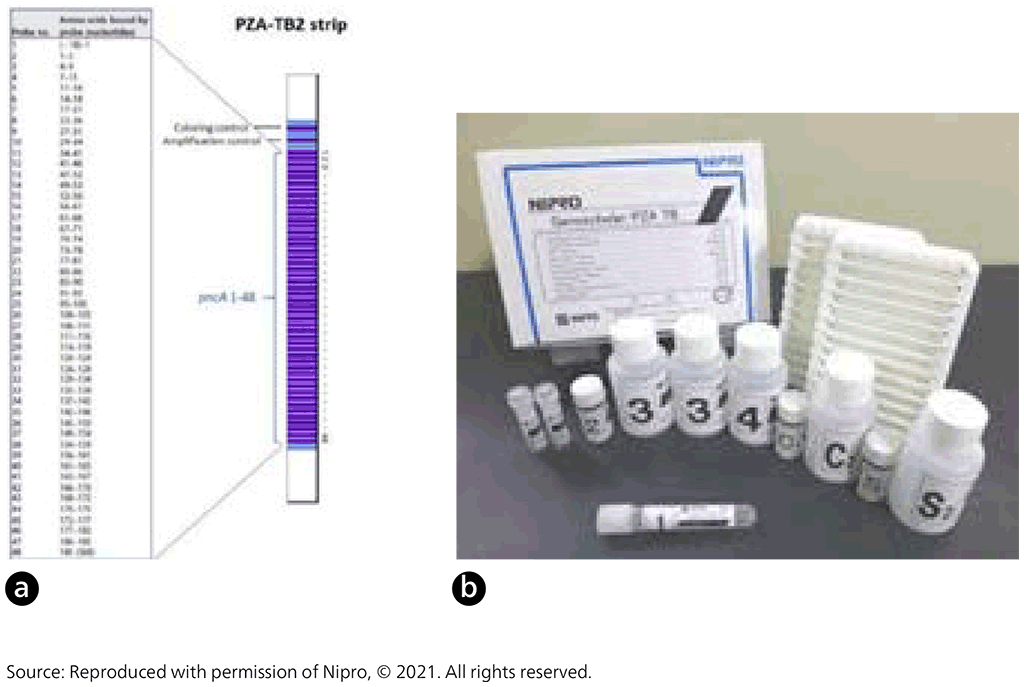
DNA extracted from cultures is amplified with primers by PCR. Amplified DNA is then hybridized to complementary oligonucleotide probes that are bound on a membrane strip. Streptavidin labelled with alkaline phosphatase is then added, to bind to any hybrids formed in the previous step. Next, a substrate is added, and an enzymatic reaction results in purple bands, which are visually interpreted. The absence of wild-type probe binding indicates the presence of a mutation. The first version of the assay contained 47 probes, which covered the pncA promoter and open reading frame. The second version contained 48 probes, three of which (pncA 16, 17 and 35) represent silent mutations known to be genetic markers not associated with pyrazinamide resistance: Gly60Gly (probe 16), Ser65Ser (probe 17) and Thr142Thr (probe 35).
Justification and evidence
The Genoscholar PZA-TB LPA assay, which is already commercially available, could potentially be implemented for diagnosis of pyrazinamide resistance in routine care. However, limited data have been published on the diagnostic accuracy of the assay. This systematic review with meta-analysis aimed to assist in collating all the available data to understand the diagnostic accuracy of the pyrazinamide LPA assay for detection of pyrazinamide resistance in TB patients, to guide policy-makers and clinicians.
The WHO Global TB Programme initiated an update of the current guidelines and commissioned a systematic review on the use of high complexity reverse hybridization-based NAATs for detection of pyrazinamide resistance in people with signs and symptoms of TB.
Two PICO questions were designed to form the basis for the evidence search, retrieval and analysis:
- Should high complexity reverse hybridization-based NAATs on sputum be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST or composite reference standard?
- Should high complexity reverse hybridization-based NAATs on isolates be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST?
The databases searched were PubMed, Web of Science and Embase, and they were searched without language or date restrictions. The search query was (PZA OR pyrazinamide OR pncA) AND (tuberculosis) AND (“line-probe assay” OR LPA OR “hybridization-based technology”). In addition, we approached Nipro (Osaka, Japan) to identify non-published data.
The microbiological reference standard was defined either as phenotypic culture-based DST performed using BD MGIT 960 PZA liquid assay or another acceptable phenotypic assay, or as genotypic DST performed using either targeted sequencing of the pncA gene or whole genome sequencing. In the case of genotypic DST, all samples with a pncA wild type were defined as being susceptible, while any variant in pncA was considered resistant, which implicitly would categorize “silent” mutations as resistant. In contrast, the composite reference standard was defined by classifying all samples with pncA wild type, pncA silent mutations and neutral mutations as being susceptible, while any other variant in pncA was considered resistant (44).
The certainty of the evidence was assessed consistently through PICO questions, using the GRADE approach (36, 37), which produces an overall quality assessment (or certainty) of evidence and a framework for translating evidence into recommendations. In the GRADE approach, even if diagnostic accuracy studies are of observational design, they start as high-quality evidence.
GRADEpro Guideline Development Tool software (20) was used to generate summary of findings tables. The quality of evidence was rated as high (not downgraded), moderate (downgraded one level), low (downgraded two levels) or very low (downgraded more than two levels), based on five factors: risk of bias, indirectness, inconsistency, imprecision and other considerations. The quality (certainty) of evidence was downgraded by one level when a serious issue was identified and by two levels when a very serious issue was identified in any of the factors used to judge the quality of evidence.
Data synthesis was structured around the two preset PICO questions, as outlined below. Three web annexes give additional information, as follows:
- details of studies included in the current analysis (Web Annex 1.9: High complexity reverse hybridization-based NAATs);
- a summary of the results and details of the evidence quality assessment (Web Annex 2.9: High complexity reverse hybridization-based NAATs); and
- a summary of the GDG panel judgements (Web Annex 3.9: High complexity reverse hybridization-based NAATs).
PICO 1: Should high complexity reverse hybridization-based NAATs on sputum be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST or composite reference standard?
Three studies with a total of 122 participants provided data for evaluation of these NAATs for detection of pyrazinamide resistance, including two studies (101 participants) with phenotypic culture-based reference standard and one study (21 participants) with genotypic reference standard. The number of studies and participants were considered insufficient to make a conclusion on a diagnostic accuracy of high complexity reverse hybridization-based NAATs on sputum.
PICO 2: Should high complexity reverse hybridization-based NAATs on isolates be used to diagnose pyrazinamide resistance in patients with microbiologically confirmed pulmonary TB, irrespective of resistance to rifampicin, as compared with culture-based phenotypic DST?
Seven studies with a total of 964 participants provided data for evaluation of these NAATs for detection of pyrazinamide resistance compared with a phenotypic culture-based reference standard (Fig. 2.3.8).
The studies suffered from selection bias because they selected isolates with a wide range of different pncA mutations rather than a representative sample from a population. Thus, the evidence was downgraded by one level for risk of bias. The included studies did not directly address the review question; hence, the evidence was downgraded one level for indirectness. The Burhan trial and the Rienthong study are outliers for their sensitivities compared with the other studies; hence, the evidence was downgraded one level for inconsistency. Taking these judgements together, the quality (certainty) of evidence was rated very low for sensitivity and low for specificity.
Fig. 2.3.8. Forest plot of included studies for pyrazinamide resistance detection, irrespective of rifampicin resistance with culture-based phenotypic DST as the reference standard
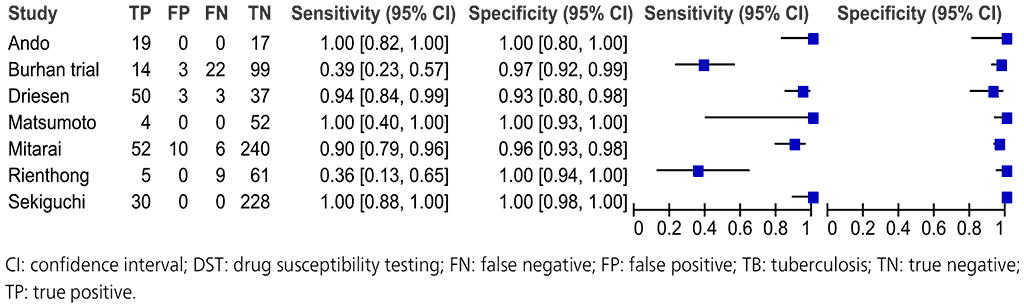
The overall sensitivity for pyrazinamide resistance in these seven studies ranged from 36% to 100% and the specificity from 96% to 100%. The pooled sensitivity was 81.2% (95% CI: 75.4–85.8%) and specificity was 97.8% (95% CI: 96.5–98.6%).
More details on diagnostic accuracy of the high complexity reverse hybridization-based NAATs, including comparison with genotypic and composite reference standards are available in Web Annex 4.17: High complexity reverse hybridization-based NAATs: diagnostic accuracy for detection of resistance to pyrazinamide. A systematic review.
Cost–effectiveness analysis
This section answers the following additional question:
What is the comparative cost, affordability and cost–effectiveness of implementation of high complexity reverse hybridization-based NAATs?
A systematic review was carried out, focusing on economic evaluations of high complexity reverse hybridization-based NAATs. Four online databases (Embase, Medline, Web of Science and Scopus) were searched for new studies published from 1 January 2010 through 17 September 2020. The citations of all eligible articles, guidelines and reviews were reviewed for additional studies. The experts and test manufacturers were also contacted to identify any additional unpublished studies.
The objective of the review was to summarize current economic evidence and further understand the costs, cost–effectiveness and affordability of high complexity reverse hybridization-based NAATs.
No published studies were identified assessing costs or cost–effectiveness using the commercially available high complexity hybridization-based NAAT (Genoscholar PZA-TB II, Nipro Japan). Indirect evidence was available from several sources. Four studies examining other commercially available LPAs (Genotype MTBDRsl and MTBDRplus, Hain Lifescience) were identified.
The Genoscholar PZA LPA was developed for use with the Nipro automated MultiBlot; however, a recent unpublished trial¹⁹ demonstrated that the Twincubator by Hain Lifescience could be used successfully with this LPA. This finding could make it easier to implement the Genoscholar PZA LPA in selected settings where Hain Lifescience equipment is already in use.
How large are the resource requirements (costs)?
No direct evidence from published studies was found regarding the total resources required. Resource requirements will include the purchase of test kits (Genoscholar PZA LPA: US$ 16/ test kit consumables only), and the equipment, which is available for US$ 14 000. Operational costs are frequently several times greater than test kit costs (and will vary across settings), but are not accounted for usually. Nipro hopes that further reductions in test costs can be achieved when the Genoscholar PZA-TB II product is distributed globally.
Unit test costs for the Genotype MTBDRsl and MTBDRplus ranged from US$ 23.46 to US$ 108.70 (45–48), with higher unit test costs in countries such as China and South Africa, largely driven by higher staff wages and operational costs. Extrapolations from unit test costs using different LPAs should be done with caution, and they are not intended to be directly transferrable estimates. Nevertheless, these indirect data do suggest that the total unit test cost of the Genoscholar PZA-TB II is likely several-fold higher than the unit test kit consumable cost of US$ 16.
Total costs will vary, depending on testing volume, numbers eligible for testing and prevalence of pyrazinamide resistance in the population. The impact on the budget will depend on the current standard of care, diagnostic and care pathways, and associated resource use.
What is the certainty of the evidence of resource requirements (costs)?
Direct costs related to test kits and machinery are available, whereas several important items related to resource use (e.g. staff time, and overhead and operational costs associated with implementing Genoscholar PZA-TB II) have not been investigated. Differences in resource use between Genoscholar PZA-TB II and existing approaches will vary across settings that are using different phenotypic and genotypic DST. Also, there is important variability in costs of staff time and operation (e.g. testing volume) across settings.
Does the cost–effectiveness of the intervention favour the intervention or the comparison?
No cost–effectiveness studies were identified using the Genoscholar PZA-TB II. Extrapolation of cost–effectiveness data from other LPAs is not advised owing to differences in diagnostic accuracy, resistance prevalence, and the testing and treatment cascade of care.
More details on economic evidence synthesis and analysis are given in Web Annex 4.9: Systematic literature review of economic evidence for NAATs to detect TB and DR-TB in adults and children.
User perspective
This section answers the following questions about key informants’ views and perspectives on the use of high complexity reverse hybridization-based NAATs:
- Is there important uncertainty about or variability in how much end-users value the main outcomes?
- What would be the impact on health equity?
- Is the intervention acceptable to key stakeholders?
- Is the intervention feasible to implement?
The synthesis and analysis of qualitative evidence on end-users’ perspectives are discussed above in the section “User perspective” for moderate complexity automated NAATs (p. 61–65).
Findings of the review and interviews
The main findings of the systematic review and interviews are given below. Where information is from the review, a level of confidence in the QES is given; where it is from interviews, this is indicated with ‘Interviews’.
Is there important uncertainty about or variability in how much end-users value the main outcomes?
- Patients in high burden TB settings value:
- getting an accurate diagnosis and reaching diagnostic closure (finally knowing “what is wrong with me”);
- avoiding diagnostic delays because they exacerbate existing financial hardships and emotional and physical suffering, and make patients feel guilty for infecting others (especially children);
- having accessible facilities; and
- reducing diagnosis-associated costs (e.g. travel, missing work) as important outcomes of the diagnostic.
QES: moderate confidence
- The high complexity reverse hybridization-based NAATs meet some preferences and values of laboratory staff and clinicians, in that the current test:
- provides quicker results about pyrazinamide resistance than other available methods (e.g. culture DST);
- can provide information on different concentration levels; and
- targets a drug that is widely used in first-line TB treatment.
Interviews
What would be the impact on health equity?
The impact on health equity would be similar to that of moderate complexity automated NAATs (p. 63–64), plus the following:
- Lengthy diagnostic delays, underuse of diagnostics, lack of TB diagnostic facilities at lower levels and too many eligibility restrictions hamper access to prompt and accurate testing and treatment, particularly for vulnerable groups.
QES: high confidence
Applicability to three index tests also confirmed in interviews - Staff and managers voiced concerns about the sustainability of funding and maintenance, complex conflicts of interest between donors and implementers, and the strategic and equitable use of resources, which makes it difficult to ensure equitable access to cartridge-based diagnostics.
QES: high confidence - For patients, access to clear, comprehensible and dependable information on what TB diagnostics are available to them and how to interpret results is a vital component of equity; lack of such access represents an important barrier for patients.
Interviews - New treatment options need to be matched with new diagnostics: it is important to improve access to treatment based on new diagnostics, and to improve access to diagnostics for new treatment options.
Interviews - The speed at which WHO guidelines are changing does not match the speed at which many country programmes are able to implement the guidelines. This translates into differential access to new TB diagnostics and treatment:
- between countries (i.e. between those that can and cannot quickly keep up with the rapidly changing TB diagnostic environment); and
- within countries (i.e. between patients who can and cannot afford the private health system that is better equipped to quickly adopt new diagnostics and policies).
Interviews
Is the intervention acceptable to key stakeholders?
- Acceptability of a high complexity reverse hybridization-based NAAT depends on how well the test performs on different samples, because laboratory staff question how well LPA methods work on smear-negative samples. If samples need to be cultured before the pyrazinamide LPA is run, this may undermine the benefits of this method’s quicker turnaround time compared with phenotypic DST for pyrazinamide. Acceptability also depends on how well the test actually detects mutations specific to pyrazinamide resistance; clinicians and laboratory staff may require further clarification and justification in some settings as to why this specific drug test is being prioritized, given that it is not currently part of routine DST.
- Specific feasibility challenges (training and infrastructure requirements, sample quality result interpretation system), general feasibility challenges (as identified in the interview study and QES, respectively) and accumulated delays risk undoing the added value and benefits identified by the users (e.g. avoiding delays and drug-resistance information).
QES high confidence and interviews
Is the intervention feasible to implement?
- The feasibility of implementing the pyrazinamide LPA is challenged by the significant training and laboratory infrastructure required to implement this method. Feasibility also hinges on the availability of an automated interpretation system, because the result is difficult to interpret.
Interviews
Implementation considerations
Factors to consider when implementing a high complexity hybridization-based NAAT for detection of pyrazinamide resistance are as follows:
- There are specific concerns about the complexity and difficulty of interpretation. The large number of bands makes it difficult to read the result of the high complexity reverse hybridization-based NAAT.
- Local epidemiological data on resistance prevalence should guide local testing algorithms, whereas pretest probability is important for the clinical interpretation of test results.
- The cost of a test varies, depending on the number of samples in a batch, staff time and other parameters requiring a local costing exercise to be performed.
- Low, moderate, and high complexity tests have a successive increase in technical competency needs (qualifications and skills) and staff time, impacting planning and budgeting.
- Availability and timeliness of local support service and maintenance should be considered when selecting a provider.
- Laboratory accreditation and compliance with a robust quality management system (including appropriate quality control) is essential for sustained service excellence and trust.
- Training of both laboratory and clinical staff will ensure effective delivery of services and clinical impact.
- Use of connectivity solutions for communication of results is encouraged, to improve efficiency of service delivery and time to treatment initiation.
- Based on a multinational, population-based study, levels of pyrazinamide resistance varied widely in the surveyed settings (3.0–42.1%). In all settings, pyrazinamide resistance was significantly associated with rifampicin resistance (49).
- Implementation of a high complexity hybridization-based NAAT requires laboratories with the required infrastructure, space and functional sample referral systems.
- Because there are several manual steps involved, well-trained staff are needed to set up assays and maintain instruments. Special training and experience are required for reading of banding patterns on the strip.
Research priorities
Research priorities for a high complexity hybridization-based NAAT for detection of pyrazinamide resistance are as follows:
- diagnostic accuracy of high complexity hybridization-based NAATs indirect testing on sputum and non-sputum samples in people with signs and symptoms of TB, with or without resistance to rifampicin;
- impact of diagnostic technologies on clinical decision-making and outcomes important to patients (e.g. cure, mortality, time to diagnosis and time to start treatment) in all patient populations;
- impact of specific mutations on treatment outcomes among people with DR-TB;
- use, integration and optimization of diagnostic technologies in the overall landscape of testing and care, as well as diagnostic pathways and algorithms;
- economic studies evaluating the costs, cost–effectiveness and cost–benefit of diagnostic technologies;
- qualitative studies evaluating equity, acceptability, feasibility and end-user values of diagnostic technologies; and
- interpretation of the results from a high complexity hybridization-based NAAT compared with sequencing and newer evidence on genotypic and phenotypic associations.
19 Leen Rigouts: Validation study of Genoscholar PZA LPA in three Supranational TB Reference Laboratories.
 Retour
Retour