Liens transversaux de livre pour First-line LPAs
In 2008, WHO approved the use of commercial LPAs for detecting MTBC in combination with resistance to rifampicin and isoniazid in sputum smear-positive specimens (direct testing) and in cultured isolates of MTBC (indirect testing). A systematic review at that time evaluated the diagnostic accuracy of two commercially available LPAs – the INNO-LiPA Rif.TB assay (Innogenetics, Ghent, Belgium), and the GenoType® MTBDRplus (version 1), hereafter referred to as Hain version 1 – and provided evidence for WHO’s endorsement (37, 38). Excellent accuracy was reported for both tests in detecting rifampicin resistance, but their diagnostic accuracy for isoniazid resistance had lower sensitivity, despite the high specificity. Because there were inadequate data to allow stratification by smear status, WHO’s recommendation for using LPAs was limited to culture isolates or smear-positive sputum specimens. Further data have since been published on the use of LPAs; newer versions of LPA technology have now been developed, such as the Hain GenoType MTBDRplus version 2, hereafter referred to as Hain version 2; and other manufacturers have entered the market, including Nipro (Tokyo, Japan), which developed the Genoscholar™ NTM+MDRTB II, hereafter referred to as Nipro.
In 2015, FIND evaluated the Nipro and the Hain version 2 LPAs, and compared them with Hain version 1. The study demonstrated equivalence among the three commercially available LPAs for detecting TB and resistance to rifampicin and isoniazid (5).
Recommendation
- For persons with a sputum smear-positive specimen or a cultured isolate of MTBC, commercial molecular LPAs may be used as the initial test instead of phenotypic culture-based DST to detect resistance to rifampicin and isoniazid.
(Conditional recommendation, moderate certainty in the evidence for the test’s accuracy)
Remarks
- These recommendations apply to the use of LPAs for testing sputum smear-positive specimens (direct testing) and cultured isolates of MTBC (indirect testing) from both pulmonary and extrapulmonary sites.
- LPAs are not recommended for the direct testing of sputum smear-negative specimens.
- These recommendations apply to the detection of MTBC and the diagnosis of MDR-TB, but acknowledge that the accuracy of detecting resistance to rifampicin and isoniazid differs and, hence, that the accuracy of a diagnosis of MDR-TB is reduced overall.
- These recommendations do not eliminate the need for conventional culture-based DST, which will be necessary to determine resistance to other anti-TB agents and to monitor the emergence of additional drug resistance.
- Conventional culture-based DST for isoniazid may still be used to evaluate patients when the LPA result does not detect isoniazid resistance. This is particularly important for populations with a high pretest probability of resistance to isoniazid.
- These recommendations apply to the use of LPA in children based on the generalization of data from adults.
Test description
LPAs are a family of DNA strip-based tests that can detect the MTBC strain and determine its drug resistance profile through the pattern of binding of amplicons (DNA amplification products) to probes targeting the following: specific parts of the MTBC genome (for MTBC detection), the most common resistance-associated mutations to first-line and second-line agents, or the corresponding wild-type DNA sequence (for detection of resistance to anti-TB drugs) (38).
LPAs are based on reverse hybridization DNA strip technology and involve three steps: DNA extraction from M. tuberculosis culture isolates or directly from patient specimens, followed by multiplex PCR amplification and then reverse hybridization with visualization of amplicon binding (or lack thereof) to wild-type and mutation probes (5).
Although LPAs are more technically complex to perform than the Xpert MTB/RIF assay, they can detect isoniazid resistance. Testing platforms have been designed for a reference laboratory setting and are thus most applicable to high TB burden countries. Results can be obtained in 5 hours.
Some of these steps can be automated, making the method quicker and more robust, and reducing the risk of contamination.
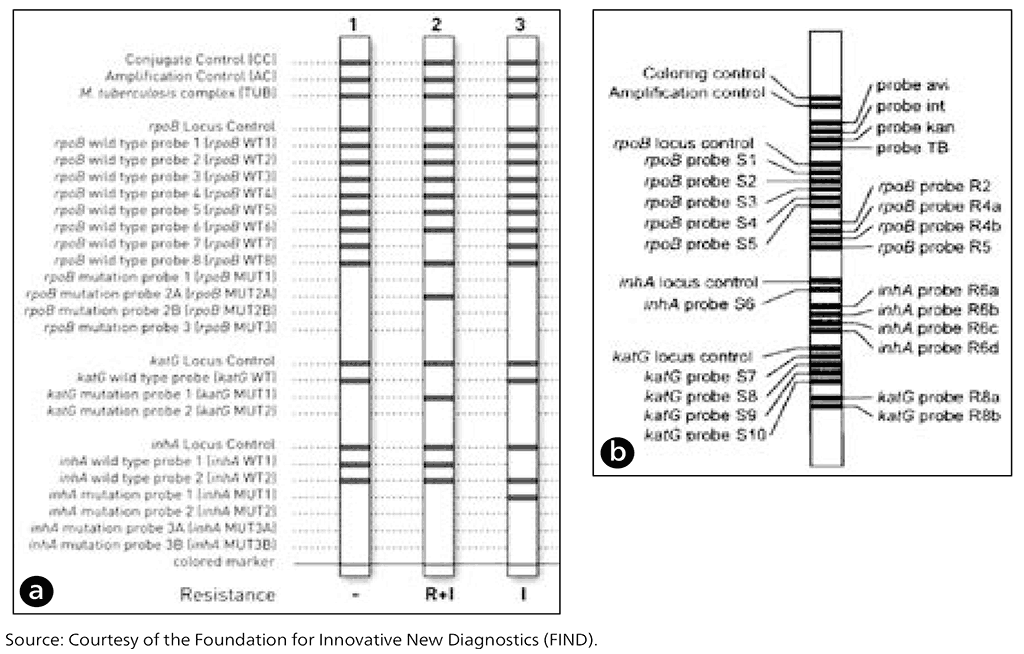
The Hain version 1 and version 2 assays include rpoB probes to detect rifampicin resistance, katG probes to detect mutations associated with high-level isoniazid resistance, and inhA promoter probes to detect mutations usually associated with low-level isoniazid resistance. The probes used to detect wild-type and specific mutations are the same for both versions of the Hain LPA (Fig. 2.3.5a).
Similarly, the Nipro assay allows for the identification of MTBC, and resistance to rifampicin and isoniazid. The Nipro assay also differentiates M. avium, M. intracellulare and M. kansasii from other non-tuberculous mycobacteria (Fig. 2.3.5b).
The rpoB, katG and inhA promoter mutation probes are the same for the three assays, with the exception of the katG S315N mutation, which is included in the Nipro assay but not in Hain version 1 or version 2. There are some minor variations in the codon regions covered for the wild type among Hain version 1 and version 2, and the Nipro.
Fig. 2.3.5. Examples of different line probe assay strip readouts: (a) Hain GenoType MTBDRplus version 1 and version 2 (Hain Lifescience, Nehren, Germany) and (b) Nipro NTM+MDRTB Detection Kit 2 (Nipro, Tokyo, Japan)
Justification and evidence
In 2015, WHO commissioned an updated systematic review of the accuracy of commercial LPAs for detecting MTBC, and resistance to rifampicin and isoniazid. A total of 74 studies were identified, comprising 94 unique datasets (see Annex 1.3: “FL-LPA”). Of these 94 datasets, 83 evaluated Hain version 1, five evaluated Hain version 2, and six evaluated the Nipro assay. Only one of the studies performed head-to-head testing of all three target LPAs on directly tested clinical specimens and indirectly tested isolates, and these data were included as six separate datasets (39). No studies performed LPA testing on specimens and culture isolates from the same patients, precluding direct within-study comparisons.
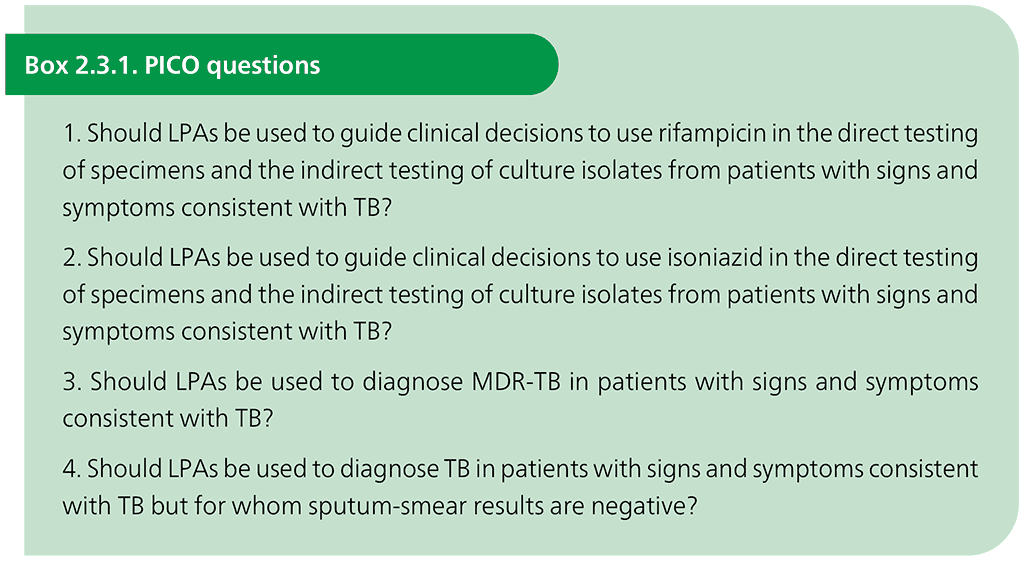
Following the 2015 systematic review, the WHO Global TB Programme convened a GDG in March 2016 to assess the data and update the 2008 policy recommendations on using commercial LPAs to detect MTBC, and resistance to isoniazid and rifampicin. The PICO questions are given in Box 2.3.1
LPAs were compared with a phenotypic culture-based DST reference standard, and a composite reference standard that combined the results from genetic sequencing with results from phenotypic culture-based DST. Phenotypic DST was the primary reference standard applied to all participants for all analyses. These analyses were stratified – first, by susceptibility or resistance to rifampicin or isoniazid (or both) and second, by type of LPA testing (indirect testing or direct testing).
Several studies contributed to either sensitivity (no true positives and no false negatives) or specificity (no true negatives and no false positives) but not to both. For these studies, a univariate, random-effects meta-analysis of the estimates of sensitivity or specificity was performed separately, to make optimal use of the data. The results from the univariate analysis (using all studies) were compared with the results from the bivariate analysis of the subset of studies that contributed to estimates of both sensitivity and specificity.
If there were at least four studies for index tests with data that contributed only to sensitivity or specificity, a univariate, random-effects meta-analysis was performed to assess one summary estimate, assuming no correlation between sensitivity and specificity. In cases in which there were fewer than four studies, or where substantial heterogeneity was evident on forest plots that precluded a meta-analysis, a descriptive analysis was performed for these index tests. Forest plots were visually assessed for heterogeneity among the studies within each index test and in the summary plots, for variability in estimates and the width of the prediction region (a wider prediction region suggests more heterogeneity).
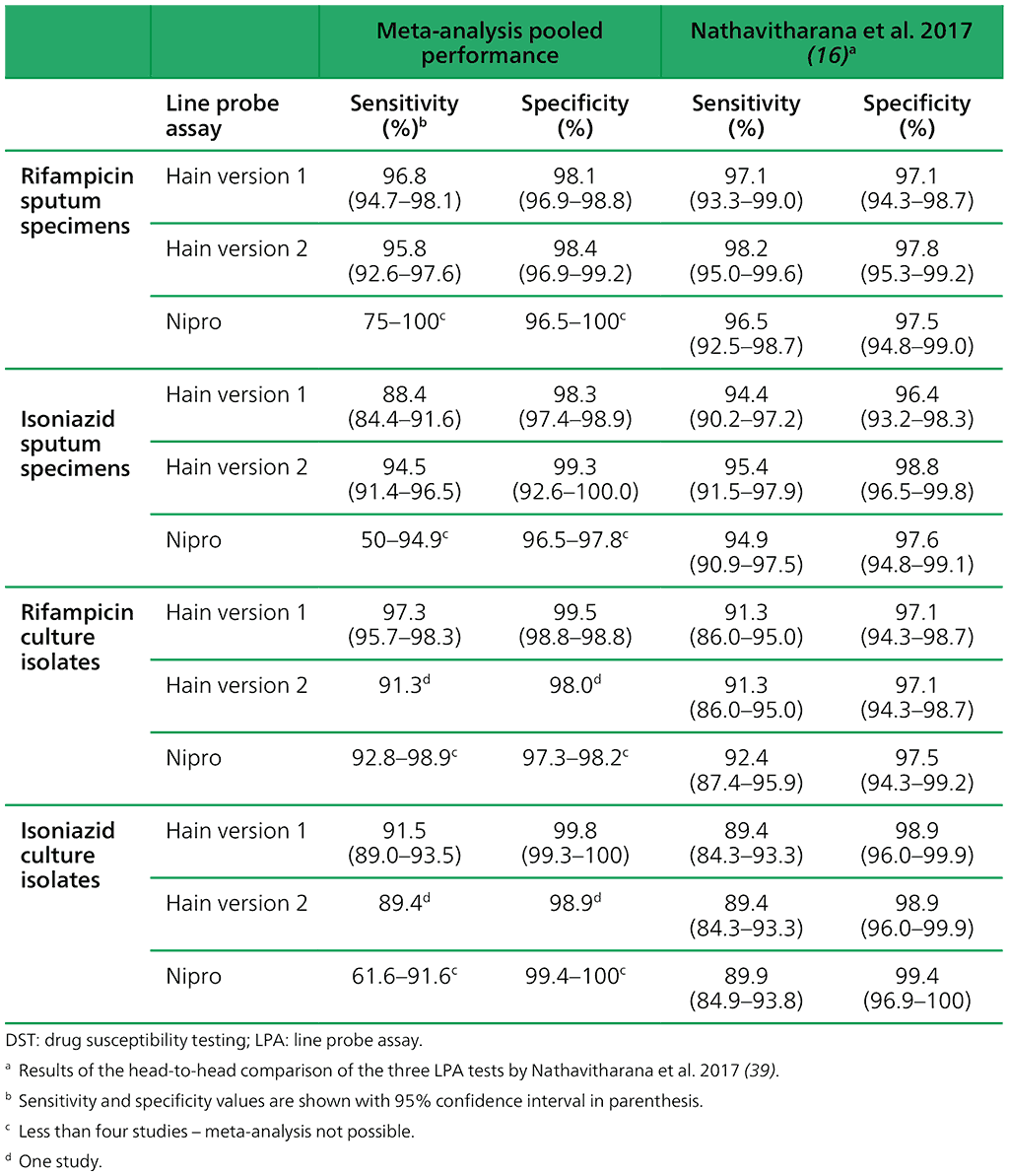
The performance of the tests is summarized in Table 2.3.2. The results are based on various numbers of studies and specimens tested. In some cases, too few studies were available for meta-analysis. The results from the only head-to-head comparison of the three tests are presented in the right-hand columns for comparison. The data presented are all comparisons with phenotypic culture-based DST as the reference standard.
Table 2.3.2. Performance of the three LPA tests for detection of rifampicin and isoniazid resistance with phenotypic culture-based DST as the reference standard
Implementation considerations
Adopting LPAs to detect rifampicin and isoniazid resistance does not eliminate the need for conventional culture and DST capacity. Culture and phenotypic culture-based DST have critical roles in monitoring patients’ responses to treatment and detecting additional resistance to second-line agents.
- The adoption of LPA should be phased in, starting at national or central reference laboratories, or those with proven capability to conduct molecular testing. Expansion could be considered, within the context of a country’s plans for laboratory strengthening, the availability of suitable personnel in peripheral centres and the quality of specimen transport systems.
- Adequate and appropriate laboratory infrastructure and equipment should be provided, to ensure that the required precautions for biosafety and the prevention of contamination are met – specimen processing for culture and procedures for manipulating cultures must be performed in biological safety cabinets in TB-containment laboratories.
- Laboratory facilities for LPAs require at least three separate rooms, one each for DNA extraction, pre-amplification procedures, and amplification and post-amplification procedures. To avoid contamination, access to molecular facilities must be restricted, a unidirectional workflow must be implemented and stringent cleaning protocols must be established.
- Appropriate laboratory staff should be trained to conduct LPA procedures. Staff should be supervised by a senior staff member with adequate training and experience in molecular assays. A programme for the external quality assessment of laboratories using LPAs should be developed as a priority.
- Mechanisms for rapidly reporting LPA results to clinicians must be established, to provide patients with the benefit of early diagnosis. The same infrastructure used for performing LPAs can be used also to perform second-line LPAs.
- LPAs are designed to detect TB and resistance to rifampicin and isoniazid in the direct testing of processed sputum samples, and in the indirect testing of culture isolates of MTBC. The use of LPAs with other respiratory samples (e.g. from bronchoalveolar lavage or gastric aspiration) or extrapulmonary samples (e.g. tissue samples, CSF or other body fluids) have not been adequately evaluated.
- The availability of second-line agents is critical in the event that resistance to rifampicin or isoniazid, or both, is detected.
- For patients with confirmed MDR/RR-TB, second-line LPAs are recommended to detect additional resistance to second-line anti-TB agents.
Research priorities
- Development of improved understanding of the correlation between the detection of resistance-conferring mutations using culture-based DST and patient outcomes.
- Review of evidence to confirm or revise different critical concentrations used in culture-based DST methods.
- Determination of the limit of detection for LPA in detecting heteroresistance.
- Determination of needs for training, assessing competency and ensuring quality assurance.
- Gathering of more evidence on the impact on mortality of initiating appropriate treatment for MDR-TB.
- Meeting the STARD for future diagnostic studies.
- Performance of country-specific cost–effectiveness and cost–benefit analyses of LPA use in different programmatic settings.
 Retour
Retour